Mod:chem3d
See also: 1C comp-lab startup,1C Timetable,Laptop use,Programs,Module 1C Script,Writing up, Don't panic.
Instructions for Use of the ChemBio3D (ChemDraw) System
The ChemBio3D/ChemDraw software can be run on the Windows Computers via invoking the Start menu (bottom left of screen) and typing ChemBio3D into the search box. The program uniquely presents both a 2D and a 3D model, with the molecule being sketched in the former and after addition of a 3rd coordinate, the 3D model in the latter. This makes it particularly easy to define quite complex organic molecules very quickly (you might note that the Avogadro program presents a different metaphor; as you draw the molecule it immediately minimises the 3D energy so that bond lengths and angles get instantly corrected).
You can download this software for installing on our own Windows or Mac computer at this site
If you need to activate the license for a pre-installed copy of ChemDraw you should also visit this site
Setting the program defaults
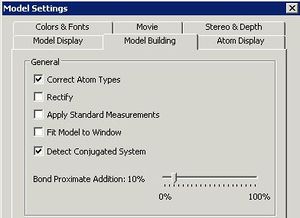
Before starting, go to File/Model Settings/Model Building. If you will be using the program for Mechanics calculation, leave the first three buttons checked (Correct building type, rectify and apply standard measurements). If you intend "importing" a file from another program as e.g. a PDB or Molfile, then deselect all these options (otherwise you may find the program eg adding hydrogens when you don't want it to). You may also find it useful to deselect these options if you are trying to rearrange the geometry of a model (the "apply standard measurements" option in particular may prevent you from moving atoms around). If in doubt, deselect all the options. Remember to "Set as default" if you want these settings to persist. Also at this stage, check the toolbars you might wish to use are displayed. There is one other important default that you will need to set relating to Gaussian calculations (see below).
Window layout
The main program display shows a 3D model on the left, a Chemdraw 2D editing pane on the right, and a results window along the bottom, together with a floating 2D editing toolbar which you can dock into the ChemDraw window if you wish. These windows can be resized as you wish.
Creating a structure using the 2D editor
Using the Chemdraw toolbar, draw the molecule in the latter as you would, with stereochemistry etc, and as you draw, the 3D equivalent will appear on the left. If you right-mouse click in the Chemdraw Window, other toolbars and options can be invoked. When finished, make sure the structure is "rectified", which is babble for "add hydrogens". First Edit/Select all, then Structure/Rectify. ChemDraw may have added the hydrogens for you already, but it often does not.
Molecular Mechanics
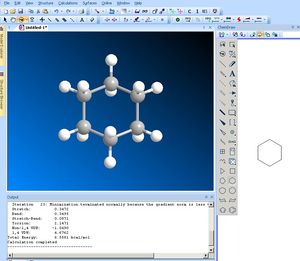
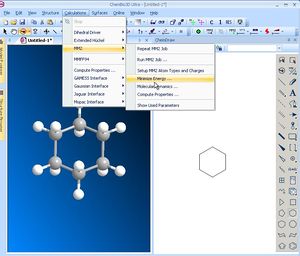
To tidy the structure up use Calculations/MM2/MinimiseEnergy then Run. When complete, the sub-division of the Mechanics energy components will be seen in the status pane at the bottom. Look particularly carefully at the components of this energy, such as the bending or stretching terms to see if you can intepret the properties of your molecule. Record the total energy (in the example here, 6.5581). An alternate force field called MMFF94 is also available (it might give slightly different results).
Rotating/Measuring/Copy-pasting
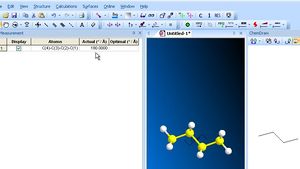
You can measure many properties in the 3D display window (right-mouse-click to see the options). The toolbar along the top of the 3D window contains a select (arrow), move (hand), rotate and zoom tool. The arrow tool along the top edge can be used to select individual atoms (and shift-select to select multiple atoms). A mouse-over the selected atoms will reveal their properties (thus a mouse-over on four atoms will reveal the dihedral angle between 1-2 and 3-4). You can also rotate the molecule using the rotation trackball tool, and move selected atoms or molecules around using the move tool at the top. To change any measurement to a value you wish it to be, select two (three or four) atoms, and from Structure/Measurement/Display Dihedral measurement edit in the new value. If instead, you type a number into the Optimal box, and re-run the MM2 calculation, it will fix this value, but optimize all the rest of the geometry. Useful if you want to obtain an approximate geometry for a transition state for example. To grab an image for your report, you can Edit/Select all the molecule, and Copy/as bitmap for pasting into eg Word.
Semi-empirical MOPAC Molecular orbital

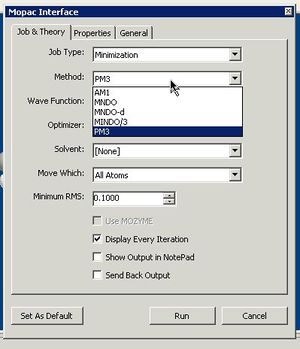

MOPAC calculations are done in a similar manner as Mechanics, but selecting Calculations/Mopac interface/Minimise(Energy/Geometry). You will then have to select the appropriate run time options (select the newest of the methods on offer, the PM6 or RM1 semi-empirical parameter set instead of the default AM1). The Results in location has to be reset to the desktop, otherwise you will get an error message from V12 of the program. This does not apply if you are running V13
Semi-empirical calculations run more slowly than the mechanics mode, but it should still be interactively fast for molecules of up to around 50 atoms. Do not attempt larger molecules unless you are prepared to wait a little time (the calculation time increases as approximately the square of the number of atoms). PM6 (but not RM1) can also be used to investigate molecules containing metals (e.g. Co etc) which cannot be run using the mechanics methods noted above.
Displaying molecular surfaces/orbitals
You can also compute properties such as Molecular surface. In order to display surface, you have to first calculate it! In the Calculations/MOPAC interface/Compute properties select Molecular Surface (assuming you have already Minimised the energy) and Run. To display the result, Surfaces/Choose Surface/Molecular Orbital will display the HOMO by default. To change the displayed orbital, Surfaces/Molecular Orbital and select the one you want if its not the HOMO. To capture the orbital for your report, Edit/select all, Edit/Copy and then in Word Edit/paste.
Ab initio/Density Functional Molecular Orbital: Batch mode

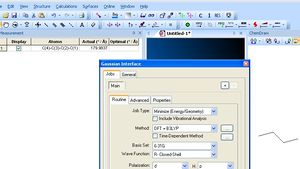
Ab initio Gaussian calculations are done in a similar manner (Calculations/Gaussian interface/Create input file). You will have to select the appropriate run time options of Method=DFT/B3LYP, 6-31G Basis set and Heavy atom d Polarisation functions and H-p functions (RB3LYP/6-31G(d,p) is the shorthand for these options as shown below and is a "standard" combination often used for research-level calculations). Save the file, adding a .gjf (or .com) suffix in your H: drive or memory stick. You will use this file (after minor editing) to submit the Gaussian job to the 'Chemistry SCAN for which detailed instructions are available here.
Ab initio/Density Functional Molecular Orbital: Interactive mode

If you don't want to wait for the SCAN to run, and for SMALL molecules only, you can run the calculation interactively, just as for MM2 and MOPAC. Be aware however that these calculations are now between 100 and 1000 times slower than e.g. MOPAC, and can take hours to finish. If you are using a HP2560p laptop, you probably do not want to have Gaussian running for more than about an hour; it might overheat! With these caveats, proceed as follows in two steps.
- Invoke Calculations/Gaussian Interface/Advanced Mode
- Type %Mem=1GB and %NProcShared=4 into the input template on separate lines
- Tune any other Gaussian keywords, ie B3LYP/6-31G(d,p) opt freq
- The Results in location has to be reset from the default to the desktop, otherwise you will get an error message from V12 of the program. This does not apply if you are running V13
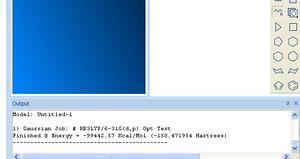
- Press Run to start the calculation. It should start a window with Gaussian 03 if you use V12 of the program (and G09 if V13). The status of the job is displayed along the bottom of the window. Note particularly the displayed Link number (L703 etc) since you can glean some idea of the progress if the link changes periodically. Gaussian spends most of its time in L501 doing the SCF, and L703 doing derivatives. It will probably cycle through e.g. L501 around 15-20 times, or more for a larger molecule. Observing this cycle will give you a very rough idea of how long one the job is going to take. If it takes more than say 10 minutes per cycle, then the total job may well run for 3 hours or so. At this stage, you might want to abandon the calculation (press on the spinning cursor in the top tool bar) and submit instead to the SCAN. The final total energy (in units of Hartree; 1 Hartree= 627.5 kcal/mol,) in this example -158.471954 should be recorded in your report. Do not record it in kcal/mol!
Ab initio/Density Functional Molecular Orbital: Displaying MOs
Molecular orbitals (and other surfaces) can be displayed in one of two ways. If you ran the Gaussian job using the SCAN, the output will be a so-called .fchk file. Double clicking this file will open Gaussview and you will use this program to inspect the MOs etc. If your molecule is small enough for Gaussian to be run interactively, proceed to display orbitals as you did with MOPAC, ie Calculation/Gaussian Interface/Compute Properties/Molecular surface and then surfaces/Choose surface/Molecular orbitals to view the HOMO, and Surfaces/Select molecular orbital to view others. Selecting, copying and pasting into a report is just as with MOPAC.
Saving the Coordinate file
When your calculation is complete, you should save the final coordinate file, via File/Save as invoking a file type of Chemical Markup language, or MDL Molfile (.mol). These files can then be used (e.g. with the Jmol applet) to incorporate into a Wiki page, or it can be read back in again to most other modelling programs (the molfile is supported by Gaussview for example).
Archiving the output into a digital repository

If you used the SCAN for your calculation, you can make use of a recent innovation; the Institutional digital repository. This is a resource for permanently archiving calculations, spectra and crystal structures. You can get a flavour of this by archiving your own calculation in the SPECTRa digital repository. To the right of the Portal display is a link termed Publish. If you click on this, and the calculation is actually in a state to be published (it may for example have failed for some reason) then appropriate metadata for the calculation are collected, and the collection deposited into the repository. From here, it can be retrieved in future.
See also: 1C comp-lab startup,1C Timetable,Laptop use,Programs,Module 1C Script,Writing up, Don't panic.
