User:Yz13712
EX3 section
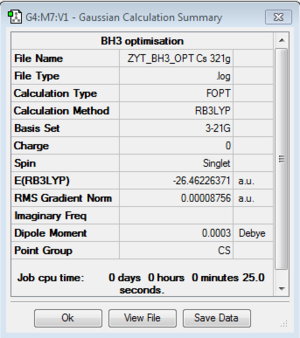
BH3:B3LYP/3-21G
Optimisation log file here
Optimisation log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| D3h | 
|
Item Value Threshold Converged? Maximum Force 0.000410 0.000450 YES RMS Force 0.000268 0.000300 YES Maximum Displacement 0.001597 0.001800 YES RMS Displacement 0.001046 0.001200 YES |
|
Optimisation log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C1 | 
|
Item Value Threshold Converged? Maximum Force 0.000228 0.000450 YES RMS Force 0.000111 0.000300 YES Maximum Displacement 0.000854 0.001800 YES RMS Displacement 0.000495 0.001200 YES |
|
BH3:B3LYP/6-31G(d,p)
A better Basis-set
Optimisation log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| Cs | 
|
Item Value Threshold Converged? Maximum Force 0.000203 0.000450 YES RMS Force 0.000098 0.000300 YES Maximum Displacement 0.000738 0.001800 YES RMS Displacement 0.000395 0.001200 YES |
|
Optimisation log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| D3h | 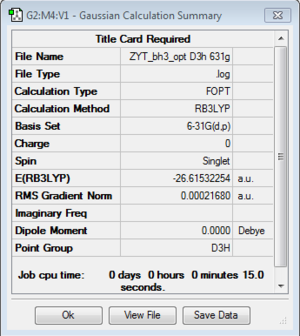
|
Item Value Threshold Converged? Maximum Force 0.000434 0.000450 YES RMS Force 0.000284 0.000300 YES Maximum Displacement 0.001702 0.001800 YES RMS Displacement 0.001114 0.001200 YES |
|
Optimisation log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C1 | 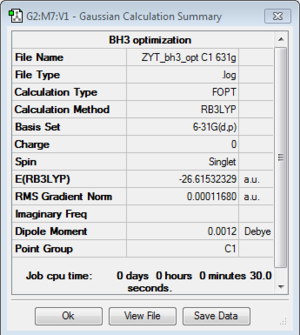
|
Item Value Threshold Converged? Maximum Force 0.000255 0.000450 YES RMS Force 0.000133 0.000300 YES Maximum Displacement 0.000920 0.001800 YES RMS Displacement 0.000598 0.001200 YES |
|
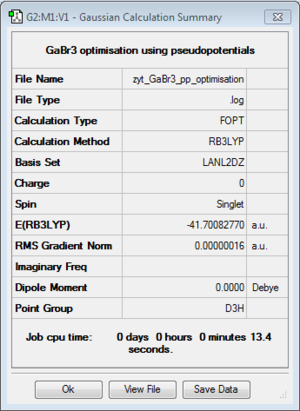
GaBr3:B3LYP/LANL2DZ
optimisation file: DOI:10042/193794
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000002 0.001200 YES |
|
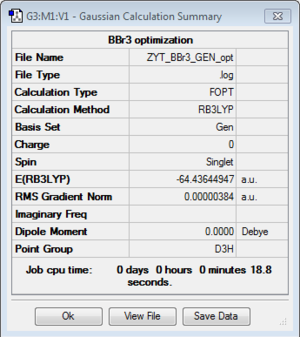
BBr3:B3LYP/6-31G(d,p)LANL2DZ (GEN)
optimisation file: DOI:10042/193796
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
|
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000036 0.001800 YES RMS Displacement 0.000024 0.001200 YES |
|
Geometry Comparison
| BH3 | BBr3 | GaBr3 | |
|---|---|---|---|
| r(E-X) Å | 1.19 | 1.93 | 2.35 |
| θ(X-E-X) degrees(º) | 120.0 | 120.0 | 120.0 |
Summary of the data: Since both central atoms (B and Ga) in EX3 system are Group 3 element, the symmetry of molecules would remain the same (point group D3h)and the bong angle is exactly 120.0º. However, the changing of ligand and central atom would affect the bond length significantly. Ga atom is much more large than B atom and hence would form a weaker bond due to the shielding effect.
Both H and Br ligands would form single bond with central atom. However the Br atom is larger than H atom, and therefore the orbital overlap for H atom is better than Br atom. Also, Br is more electronegative than H. Hence the bond length of E-Br would be larger than bond length of E-H.
Bond represents the electronic interaction between atoms. The MO theory suggest that electrons are not related to any individual atoms but will move around the whole molecule. The strength of the bond decrease along with the increase of bond length. An ionic bond is generally considered as a strong bond. for example, the ionic bond energy for NaCl is −756 kJ/mol. The single covalent bonds formed by elements from first three row of the periodic table are generally considered as medium bonds. For example, C-C bond has a bond enthalpy of 348 KJ/mol. Hydrogen bonds and other intermolecular interactions (e.g. Van Der Waals force) are often considered as weak bonds and the hydrogen bond is stronger than other intermolecular interactions. The strength of hydrogen bond in H3O is 22 KJ/mol.
The GaussView could only recognize a bond if the distance between the two atoms is smaller than the pre-set value. These values are based on organic molecules, however inorganic molecules will often have longer bonds.
Frequency Analysis
BH3:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
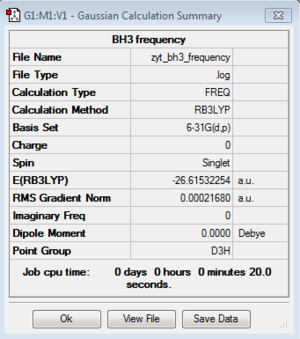
Energy exactly the same as previous optimization 6-31G BH3 model. |
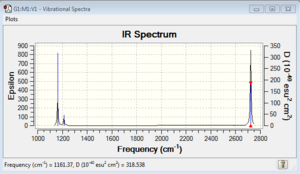
Low frequencies --- -7.0630 -7.0272 -0.0278 -0.0010 0.7100 6.6480 Low frequencies --- 1163.0024 1213.1577 1213.1579 |
Vibrational spectrum for BH3
| wavenumber | Intensity | IR active? | type |
| 1161 | 93 | yes | bend |
| 1212 | 14 | very slight | bend |
| 1212 | 14 | very slight | bend |
| 2588 | 0 | no | stretch |
| 2721 | 126 | yes | stretch |
| 2721 | 126 | yes | stretch |
There are two reasons for why there are fewer vibrational peaks than there are vibrations. The first reason is that some vibration modes does not involve in the change of dipole moment and therefore is IR inactive, for example, the stretch vibrational mode at 2582 cm-1 is IR inactive. The second reason is some of vibrations are degenerated and have exactly the same energy level like two vibrational modes at 1212 cm-1 would only show one peak at IR spectrum.
GaBr3:B3LYP/LANL2DZ
Frequency file: DOI:10042/193819
| summary data | low modes |
|---|---|
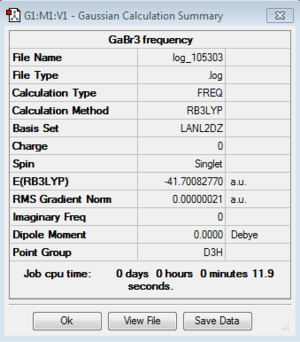
Energy exactly the same as previous optimization GaBr3 model. |
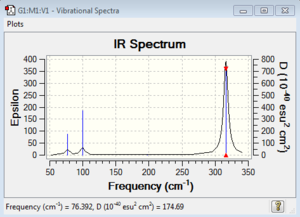
Low frequencies --- -1.4877 -0.0015 -0.0002 0.0096 0.6540 0.6540 Low frequencies --- 76.3920 76.3924 99.6767 |
Vibrational spectrum for GaBr3
| wavenumber | Intensity | IR active? | type |
| 76 | 3 | very slight | bend |
| 76 | 3 | very slight | bend |
| 100 | 9 | very slight | bend |
| 197 | 0 | no | stretch |
| 316 | 57 | yes | stretch |
| 316 | 57 | yes | stretch |
Compare and Contrast:
Large difference in vibrational frequencies
Since both Ga and Br atoms are larger than B and H atoms indicate a poor orbital overlap and weaker bond between Ga-Br than B-H. Therefore the energy required for stretch or bend the GaBr3 molecule will be much lower than BH3 molecules. Hence the vibrational frequencies of the GaBr3 molecules are much lower than those of the BH3 molecules.
Reordering of the modes
By looking at the vibrational modes of GaBr3 and BH3 in GaussView, both of two molecules contain two types of vibrational modes: one is the umbrella bending motion ( 1163 cm-1 for BH3 and 100 cm-1 for GaBr3) and another one is a pair of equivalent twisted scissoring bending motions( 1213 cm-1 for BH3 and 76 cm-1 for GaBr3). The umbrella motion requires out of plane vibrations of either the central atom or the terminal atoms, where as the twisted scissoring motion requires in-plane vibration of both central and terminal atoms
For BH3, the H atoms are much lighter than the central B atom, therefore, vibrating all of the H atoms requires less energy than vibrating central B atoms and two of the terminal H atoms, which explain the reason why umbrella motion is at a lower frequency.
For GaBr3, the central Ga atom is only slightly lighter than the terminal Br atoms, which makes the umbrella motion less favorable due to the requirement of large change in momentum. Therefore, the umbrella motion for GaBr3 is at a higher frequency.
Form GaussView, we could also see that the umbrella motion for BH3 is dominated by H atoms whereas for GaBr3 is dominated by central Ga atom.
Answer to the questions:
The computation method and basis set determine the Potential Energy Surface generated for a molecule. The geometry optimisation is based on the first derivative which locates the minimum on the potential energy surface, where as the frequency calculation is based on the second derivative around the minimum point. If the optimization and frequency analysis are calculated based on different surface, then they are not directly related and there is no point of comparing them.
Frequency analysis (second derivative) can be used to determine whether the stationary point found by the optimization is a minimum or not. If there are any negative frequencies appearing, it means that the molecule is at a saddle point or transition state rather than ground state.
Low frequencies represent the translational and rotational motions of the center of mass of the molecule. Ideally we want the low frequencies to be zero due to the Born-Oppenheimer approximation being made in these calculations, in which we assume the nucleus to be stationary comparing to electrons.
Molecular Orbitals for BH3
MO file: DOI:10042/193820
There are no significant differences between the real and LCAO MOs, this indicate that the qualitative MO theory is quite fit in predicting the general shapes of the MOs of small molecules.
NH3:B3LYP/6-31G(d,p)
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C3v | 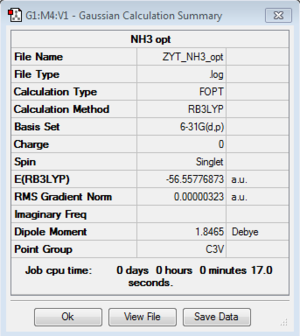
|
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000012 0.001800 YES RMS Displacement 0.000008 0.001200 YES |
|
NH3:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
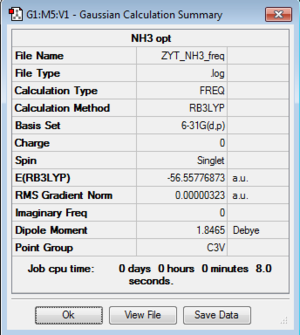
Energy exactly the same as previous optimization 6-31G BH3 model. |
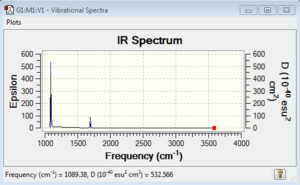
Low frequencies --- -0.0130 -0.0029 -0.0019 7.1032 8.1046 8.1049 Low frequencies --- 1089.3834 1693.9368 1693.9368 |
Vibrational spectrum for NH3
| wavenumber | Intensity | IR active? | type |
| 1089 | 145 | yes | bend |
| 1694 | 14 | slight | bend |
| 1694 | 14 | slight | bend |
| 3461 | 1 | no | stretch |
| 3590 | 0 | No | stretch |
| 3590 | 0 | No | stretch |
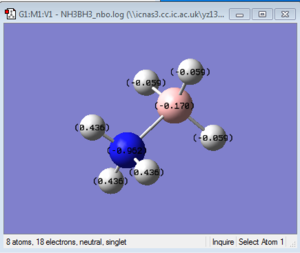
NBO Analysis of NH3
NBO log file: here
charge range: from -1.0 to 1.0.
Association Energy for NH3BH3
NH3BH3:B3LYP/6-31G(d,p)
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C3v | 
|
Item Value Threshold Converged? Maximum Force 0.000122 0.000450 YES RMS Force 0.000058 0.000300 YES Maximum Displacement 0.000531 0.001800 YES RMS Displacement 0.000296 0.001200 YES |
|
NH3BH3:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
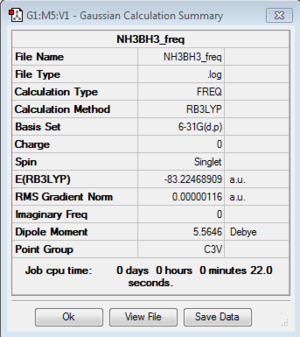
Energy exactly the same as previous optimization 6-31G NH3BH3 model. |
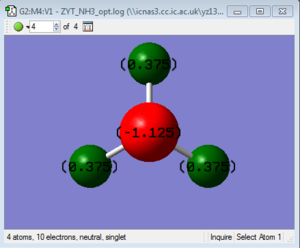
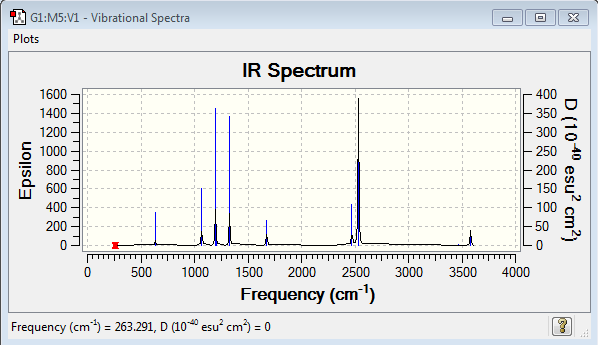
Low frequencies --- -5.4077 -0.3705 -0.0565 0.0002 0.9580 1.0693 Low frequencies --- 263.2907 632.9543 638.4541 |
Vibrational spectrum for NH3BH3
| wavenumber (cm-1) | Intensity | IR active? | type |
| 263 | 0 | no | bend |
| 633 | 14 | slight | strecth |
| 638 | 4 | very slight | bend |
| 638 | 4 | very slight | bend |
| 1069 | 41 | yes | bend |
| 1069 | 41 | yes | bend |
| 1196 | 108 | yes | bend |
| 1204 | 3 | very slight | bend |
| 1204 | 3 | very slight | bend |
| 1329 | 114 | yes | bend |
| 1676 | 28 | slight | bend |
| 1676 | 28 | slight | bend |
| 2472 | 67 | yes | stretch |
| 2532 | 231 | yes | stretch |
| 2532 | 231 | yes | stretch |
| 3464 | 3 | very slight | stretch |
| 3581 | 28 | slight | stretch |
| 3581 | 28 | slight | stretch |
Association energy calculation
E(NH3)=-56.55776873 A.U.
E(BH3)=-26.61532254 A.U.
E(NH3BH3)=-83.22468891 A.U.
ΔE=ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]=-83.22468891 A.U - [-56.55776873 A.U. + -26.61532254 A.U.]= -0.05159746 A.U. =-135 KJ/mol
The disassociation energy of NH3BH3 is obviously greater than hydrogen bonds (22 KJ/mol)and intermolecular attractions, however, it is still considerably smaller than most of the single covalent bond. For example, the B-N bond enthalpy is smaller than typical C-C bond enthalpy(348 KJ/mol), but nearly the same as a N-N single bond (170KJ/mol). Therefore, the B-N bond in NH3BH3 could be more like a medium bond.
NBO Analysis of ethane and aminoborane
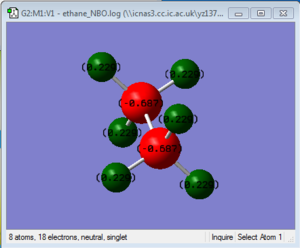
Ethane NBO log file: here
charge range: from -0.687 to 0.687.
Aminoborane NBO log file: here
charge range: from -0.962 to 0.962.
In ethane, the charge distribution for both C atoms and H atoms are equal(both C atoms is -0.687 and H atoms is 0.220). However, for aminoborane, the charge distribution for two parts are no longer the same: N atom is -0.962, B atom is -0.170 and H atoms net to N is 0.436, H atom next to B is -0.059. These all indicate that the bonds in aminoborane is highly polarized. This fits the character of the aminoborane dative-bond that N atom itself "provides" two electron to B atom. To weaken the N-B bond, we only need to substitute H atoms next to N atom into some electron withdrawing groups (e.g. -OH group) or substitute H atoms next to B atom into some electron donating groups (e.g. -CH3 group).
Ionic Liquid Project: Part 1
[N(CH3)4]+:B3LYP/6-31G(d,p)
Optimization of [N(CH3)4]+
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| TD | 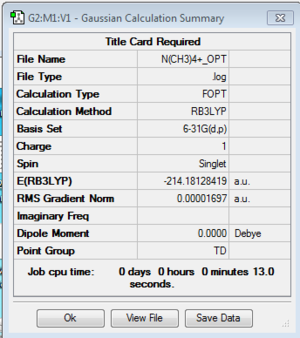
|
Item Value Threshold Converged? Maximum Force 0.000018 0.000450 YES RMS Force 0.000010 0.000300 YES Maximum Displacement 0.000231 0.001800 YES RMS Displacement 0.000089 0.001200 YES |
|
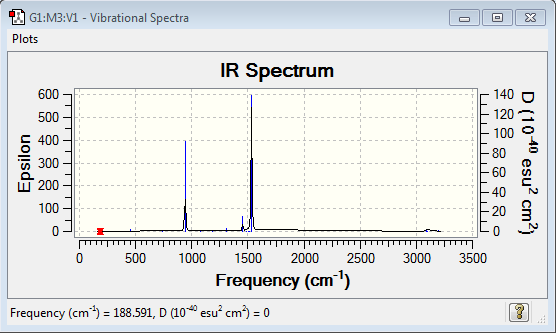
Frequency of [N(CH3)4]+:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
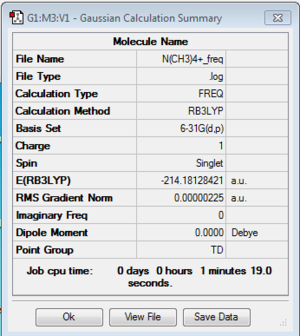
|
Low frequencies --- -0.0006 -0.0002 0.0007 21.4457 21.4458 21.4458 Low frequencies --- 188.5907 292.6934 292.6934 |
Vibrational spectrum for [N(CH3)4]+
| wavenumber (cm-1) | Intensity | IR active? | type |
| 941 | 22 | yes | bend |
| 941 | 22 | yes | bend |
| 941 | 22 | yes | bend |
| 1305 | 1 | very slight | bend |
| 1305 | 1 | very slight | bend |
| 1305 | 1 | very slight | bend |
| 1456 | 5 | slight | bend |
| 1456 | 5 | slight | bend |
| 1456 | 5 | slight | bend |
| 1532 | 53 | yes | bend |
| 1532 | 53 | yes | bend |
| 1532 | 53 | yes | bend |
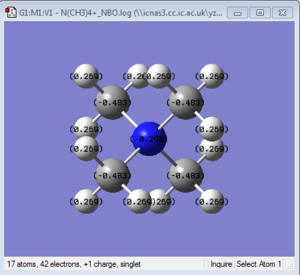
NBO Analysis of [N(CH3)4]+
NBO log file: here
charge range: from -0.483 to 0.483.
[P(CH3)4]+:B3LYP/6-31G(d,p)
Optimization of [P(CH3)4]+
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| TD | 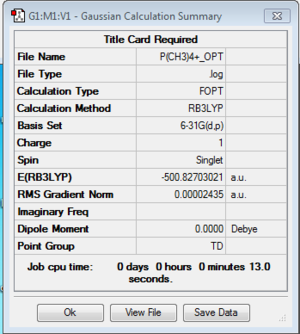
|
Item Value Threshold Converged? Maximum Force 0.000116 0.000450 YES RMS Force 0.000033 0.000300 YES Maximum Displacement 0.000809 0.001800 YES RMS Displacement 0.000326 0.001200 YES |
|
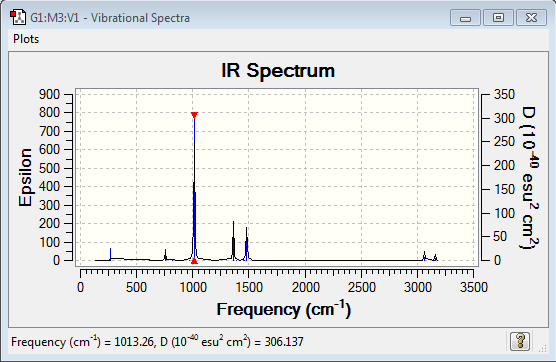
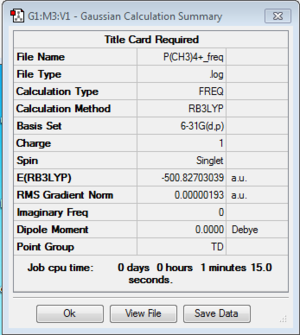
Frequency of [P(CH3)4]+:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
|
Low frequencies --- -0.0022 -0.0012 0.0022 24.7170 24.7170 24.7170 Low frequencies --- 160.1015 194.8298 194.8298 |
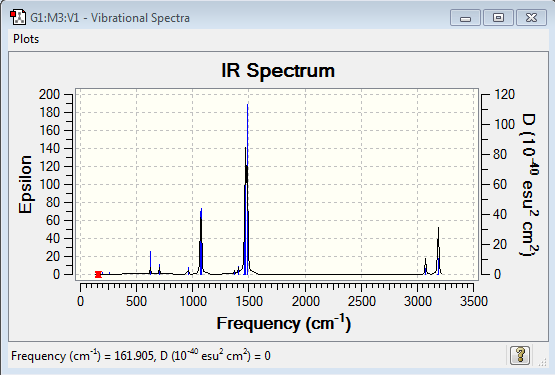
Vibrational spectrum for [P(CH3)4]+
| wavenumber (cm-1) | Intensity | IR active? | type |
| 271 | 2 | very slight | bend |
| 271 | 2 | very slight | bend |
| 271 | 2 | very slight | bend |
| 756 | 4 | slight | bend |
| 756 | 4 | slight | bend |
| 756 | 4 | slight | bend |
| 1013 | 78 | yes | bend |
| 1013 | 78 | yes | bend |
| 1013 | 78 | yes | bend |
| 1362 | 21 | yes | bend |
| 1362 | 21 | yes | bend |
| 1362 | 21 | yes | bend |
| 1481 | 26 | yes | bend |
| 1481 | 26 | yes | bend |
| 1481 | 26 | yes | bend |
| 3064 | 5 | slight | stretch |
| 3064 | 5 | slight | stretch |
| 3064 | 5 | slight | stretch |
| 3159 | 4 | slight | stretch |
| 3159 | 4 | slight | stretch |
| 3159 | 4 | slight | stretch |
NBO Analysis of [P(CH3)4]+
NBO log file: here
charge range: from -1.667 to 1.667.
[S(CH3)3]+:B3LYP/6-31G(d,p)
Optimization of [S(CH3)3]+
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C3v | 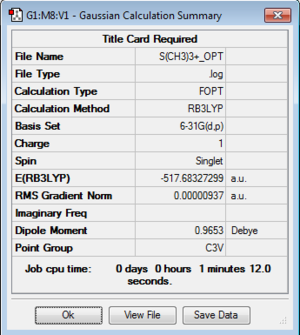
|
Item Value Threshold Converged? Maximum Force 0.000027 0.000450 YES RMS Force 0.000008 0.000300 YES Maximum Displacement 0.000235 0.001800 YES RMS Displacement 0.000073 0.001200 YES |
|
Frequency of [S(CH3)3]+:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
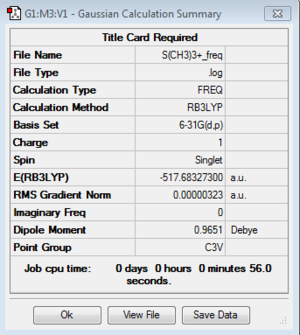
|
Low frequencies --- -7.9007 -7.6403 -7.6370 0.0019 0.0040 0.0093 Low frequencies --- 161.9049 199.6302 199.6302 |
Vibrational spectrum for [S(CH3)3]+
| wavenumber (cm-1) | Intensity | IR active? | type |
| 624 | 2 | very slight | bend |
| 704 | 1 | very slight | bend |
| 704 | 1 | very slight | bend |
| 958 | 1 | very slight | bend |
| 958 | 1 | very slight | bend |
| 1071 | 11 | yes | bend |
| 1071 | 11 | yes | bend |
| 1076 | 12 | yes | bend |
| 1408 | 2 | very slight | bend |
| 1464 | 10 | yes | bend |
| 1464 | 10 | yes | bend |
| 1473 | 25 | yes | bend |
| 1473 | 25 | yes | bend |
| 1485 | 42 | yes | bend |
| 3075 | 3 | very slight | stretch |
| 3075 | 3 | very slight | stretch |
| 3185 | 8 | slight | stretch |
| 3185 | 8 | slight | stretch |
| 3187 | 2 | very slight | stretch |
| 3187 | 2 | very slight | stretch |
| 3187 | 2 | very slight | stretch |
NBO Analysis of [S(CH3)3]+
NBO log file: here
charge range: from -0.917 to 0.917.
Geometry Comparison of [X(CH3)y]+ molecules
| [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)3]+ | |
|---|---|---|---|
| r(X-C) Å | 1.51 | 1.82 | 1.82 |
| r(C-H) Å | 1.09 | 1.09 | 1.09 |
| θ(C-X-C) degrees(º) | 109.5 | 109.5 | 102.7 |
| θ(H-C-H) degrees(º) | 110.0 | 109.0 | 109.4 ; 111.1 |
| θ(X-C-H) degrees(º) | 108.9 | 109.9 | 107.3 ; 110.6 |
The bond length of C-N is lower than C-P and C-s bond length. This is because both C and N are in the second row of Periodic Table whereas P and S are in the third row. Therefore the C and N atom will have better orbital overlap than C with P or S atom. The bond length between C-H bond did not changed much between three molecules (all are 1.09Å) which means that the change of central atom could not have too much influence on the strength of C-H bond.
The geometry shape of [N(CH3)4]+ molecule and [P(CH3)4]+ molecule are similar for they both have Td symmetry. The 4 methyl groups are all symmetrical due to the ideal bond angle (109.5º) between carbon and central atom. However, the electron distribution of P atom is larger and more diffusive than N atom and therefore would slightly compress the electrons in methyl groups results in lower H-C-H bond angle.
The [S(CH3)3]+ molecule has C3V symmetry and has two different values of H-related bond angle. This is because the total 9 H atoms are in 2 different environment: 6 of them are pointing upward to the central atom and the other 3 atoms (labelled) are pointing downward like the graph below has shown. The lone-pair electrons in S atom will push the 3 methyl groups together results in a significant decrease in C-X-C bond angle.
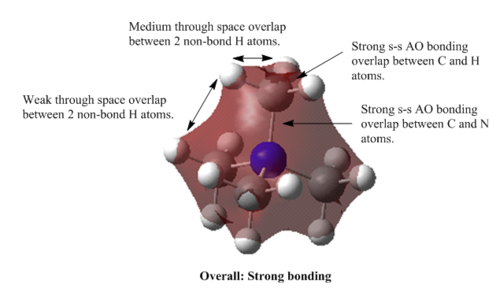
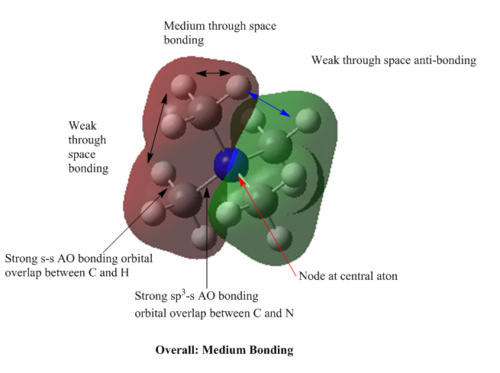
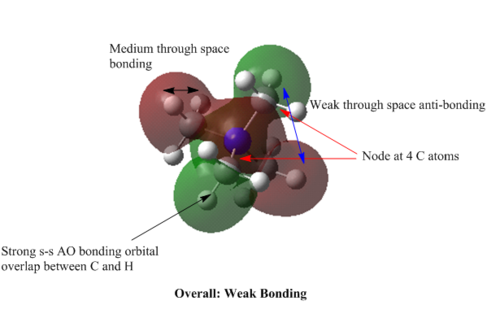
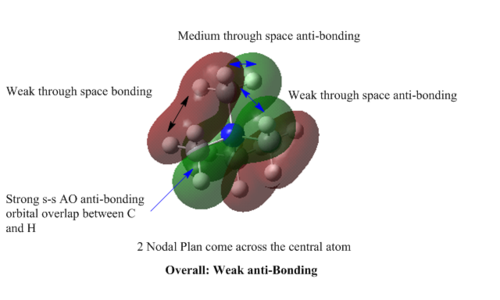
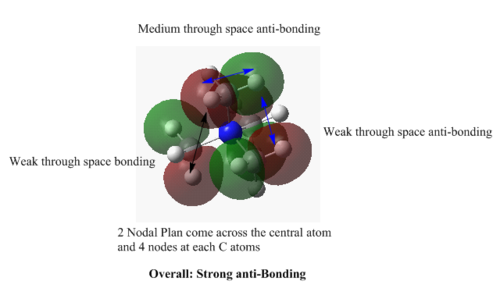
MOs of [N(CH3)4]+
MO=6 : strong bonding
MO=7 : medium bonding
MO=11 : weak bonding
MO=15 : weak anti-bonding
MO=16 : strong anti-bonding
[X(CH3)y]+ Charge Distribution (NBO) Comparison
| [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)3]+ | |
|---|---|---|---|
| charge on X | -0.295 | 1.667 | 0.917 |
| charge on C | -0.483 | -1.060 | -0.846 |
| charge on H | 0.269 | 0.298 | 0.297 ; 0.279 |
The charge on N atom is negative value whereas charges on S and P is positive values and P atom has higher value. This is due to the electronegativity among this three atoms: N has the highest electronegativity value which means that N atom is the most powerful to attract electrons therefore has negative charge value. P atom, on the contrary, has lowest electronegativity value and gets highest positive charge value.
The values of charge on C atoms is closely related to the electronegativity of their neighboring central atoms. The higher the electronegativity of their neighboring central atom is, the lower charge value on C atom will get. Therefore, Carbon next to N atom has lowest negative charge value and Carbon next to P atom has highest negative charge value. The reason why the charge on C among all three molecules are negative is because they are all methyl group and Hydrogens next to C is very electropositive.
The charges on H nearly remains the same reflect that the central atom is too far away that could not have significant affect to Hydrogen atoms. The reason why there are two different Hydrogen environments on [S(CH3)3]+ molecule has already been discussed above.
[X(CH3)y]+ Relative Contribution form C and X to the C-X Bond Comparison
| [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)3]+ | |
|---|---|---|---|
| Contribution from X | 66% | 40% | 51% |
| Hybridisation of X orbital | 25% s + 75% p | 25% s + 74% p + 1% d | 17% s + 82% p + 1% d |
| Contribution from C | 34% | 60% | 49% |
| Hybridisation of C orbital | 21% s + 79% p | 25% s + 75% p | 20% s + 80% p |
The C-X Bond interaction between all three molecules are roughly SP3 hybridization. This explain why Hyridization of X central orbitals are around 25% s + 75% p. P and S orbital contains a little d orbital contribution is because they are in the third row of Periodic Table. The hybridization of C atoms are also SP3 hybridization. Since C atom is connect to 3 H atoms and 1 X atom, the contribution of C orbital will be not exact to 25% s + 75% p.
The contribution from C to X is also strongly related to the electronegativities of X atoms. X atom with highest electronegativity (N atom) will also has the highest contribution since it attract electrons strongly.
Validity of the traditional [N(R)4]+ description
The "formal" positive charge on the N in the traditional picture represents that the positive charge is on central N atom since N atom has "donated" one electron to an Lewis base to maintain its stable 8-electron structure and SP3 Hybridization.
However, if we look at the electronegativity of each atoms inside the molecule, we will see that the positive charge is well separated among the whole structure instead of fix at any atom position.
Ionic Liquid Project: Part 2
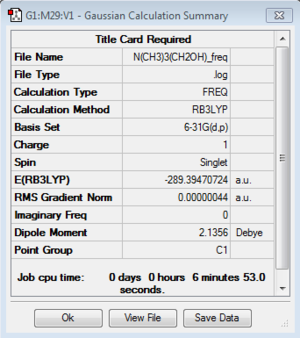
[N(CH3)3(CH2OH)]+:B3LYP/6-31G(d,p)
Optimization of [N(CH3)3(CH2OH)]+
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| C1 | 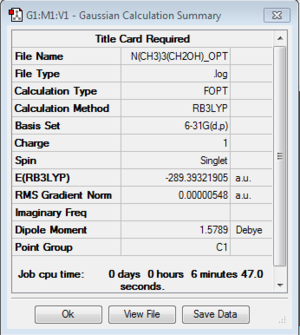
|
Item Value Threshold Converged? Maximum Force 0.000015 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000428 0.001800 YES RMS Displacement 0.000112 0.001200 YES |
|
Frequency of [N(CH3)3(CH2OH)]+:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
|
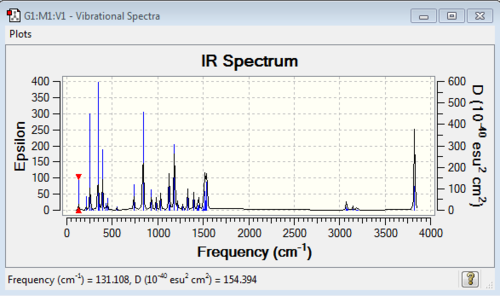
Low frequencies --- -8.4390 -5.0098 -1.0369 -0.0005 -0.0004 0.0009 Low frequencies --- 131.1078 213.4644 255.7166 |
Vibrational spectrum for [N(CH3)3(CH2OH)]+
| wavenumber (cm-1) | Intensity | IR active? | type |
| 131 | 5 | slight | bend |
| 213 | 3 | very slight | bend |
| 256 | 29 | yes | bend |
| 267 | 1 | very slight | bend |
| 342 | 51 | yes | bend |
| 355 | 4 | very slight | bend |
| 393 | 28 | yes | bend |
| 434 | 4 | very slight | bend |
| 449 | 4 | very slight | bend |
| 552 | 2 | very slight | bend |
| 736 | 22 | yes | bend |
| 839 | 96 | yes | bend |
| 931 | 22 | yes | bend |
| 982 | 12 | yes | bend |
| 1033 | 20 | yes | bend |
| 1122 | 38 | yes | bend |
| 1133 | 7 | slight | bend |
| 1184 | 91 | yes | bend |
| 1219 | 8 | slight | bend |
| 1276 | 6 | slight | bend |
| 1289 | 2 | very slight | bend |
| 1330 | 19 | yes | bend |
| 1397 | 17 | yes | bend |
| 1433 | 3 | very slight | bend |
| 1445 | 7 | slight | bend |
| 1452 | 9 | slight | bend |
| 1496 | 5 | slight | bend |
| 1501 | 3 | very slight | bend |
| 1504 | 1 | very slight | bend |
| 1514 | 26 | yes | bend |
| 1521 | 33 | yes | bend |
| 1530 | 17 | yes | bend |
| 1540 | 51 | yes | bend |
| 3074 | 9 | slight | stretch |
| 3085 | 2 | very slight | stretch |
| 3088 | 2 | very slight | stretch |
| 3095 | 1 | very slight | stretch |
| 3147 | 4 | very slight | stretch |
| 3184 | 1 | very slight | stretch |
| 3189 | 1 | very slight | stretch |
| 3825 | 105 | yes | stretch |
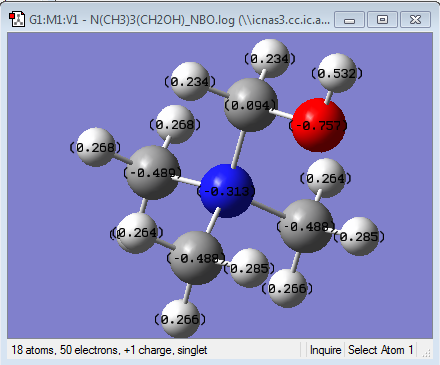
NBO analysis for [N(CH3)3(CH2OH)]+
Population log file here
charge range -0.757 to 0.757
[N(CH3)3(CH2CN)]+:B3LYP/6-31G(d,p) level
Optimization of [N(CH3)3(CH2CN)]+
Optimization log file here
| geometry | summary data | convergence | Jmol | |||
|---|---|---|---|---|---|---|
| Cs | 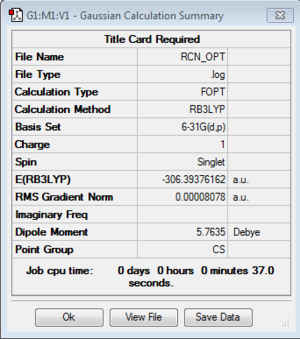
|
Item Value Threshold Converged? Maximum Force 0.000250 0.000450 YES RMS Force 0.000057 0.000300 YES Maximum Displacement 0.001338 0.001800 YES RMS Displacement 0.000490 0.001200 YES |
|
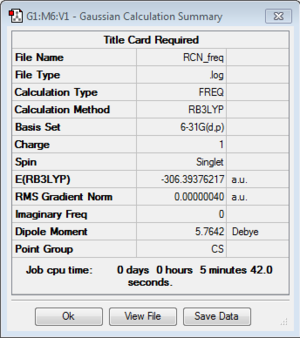
Frequency of [N(CH3)3(CH2CN)]+:B3LYP/6-31G(d,p)
Frequency file: here
| summary data | low modes |
|---|---|
|
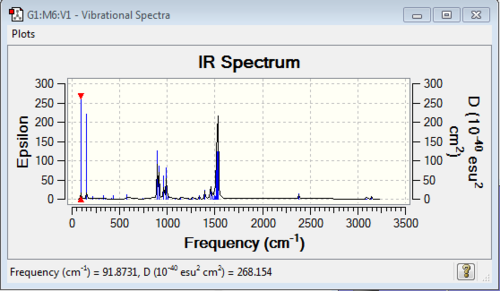
Low frequencies --- -3.6828 0.0004 0.0005 0.0011 2.7427 3.8837 Low frequencies --- 91.8731 153.9653 212.1350 |
Vibrational spectrum for [N(CH3)3(CH2CN)]+
| wavenumber (cm-1) | Intensity | IR active? | type |
| 92 | 6 | slight | bend |
| 154 | 9 | slight | bend |
| 328 | 1 | very slight | bend |
| 436 | 1 | very slight | bend |
| 571 | 2 | very slight | bend |
| 895 | 28 | yes | bend |
| 911 | 20 | yes | bend |
| 963 | 14 | yes | bend |
| 990 | 20 | yes | bend |
| 1008 | 2 | very slight | bend |
| 1140 | 1 | very slight | bend |
| 1259 | 1 | very slight | bend |
| 1333 | 3 | very slight | bend |
| 1395 | 8 | slight | bend |
| 1454 | 8 | slight | bend |
| 1455 | 8 | slight | bend |
| 1475 | 3 | very slight | bend |
| 1495 | 3 | very slight | bend |
| 1503 | 3 | very slight | bend |
| 1519 | 34 | yes | bend |
| 1521 | 47 | yes | bend |
| 1532 | 61 | yes | bend |
| 2385 | 8 | slight | stretch |
| 3087 | 1 | very slight | stretch |
| 3144 | 2 | very slight | stretch |
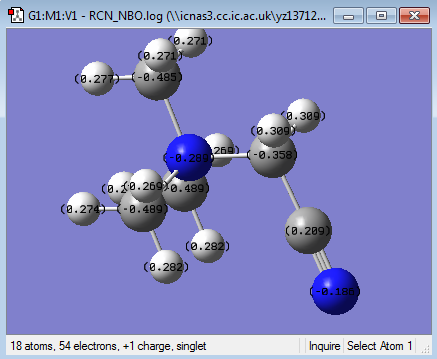
NBO analysis for [N(CH3)3(CH2CN)]+
Population log file here
charge range -0.489 to 0.489
Charge distribution comparison
| [N(CH3)3(CH2OH)]+ | [N(CH3)3(CH2CN)]+ | [N(CH3)4]+ | |
|---|---|---|---|
| charge on N | -0.313(central atom) | -0.289(central atom), -0.186(CN) | -0.295 (central atom) |
| charge on C | -0.488, -0.489(CH3), 0.094(CH2OH) | -0.489(CH3),-0.385 (the CH2 next to CN), 0.209((CN) | -0.483 (CH3) |
| charge on O | -0.757 | N/A | N/A |
| charge on H | 0.264 to 0.285(CH3), 0.234(CH2OH), 0.532(CH2OH) | From 0.269 to 0.309 | 0.269 |
Compare to the charge distribution of [N(CH3)4]+ molecule, OH group is an electron donating group and push the electrons towards the central atom which results in the increase charge value of Central N atom and other C and H atoms in the system. On the contrary, CN group is an electron withdrawing group which pull electrons from the central atom and results in the decrease value of N central atom.
HOMO and LUMO comparison
| [N(CH3)3(CH2OH)]+ | [N(CH3)3(CH2CN)]+ | [N(CH3)4]+ | |
|---|---|---|---|
| HOMO | 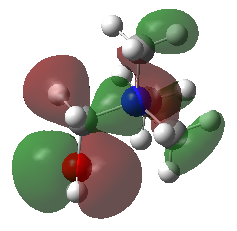 |
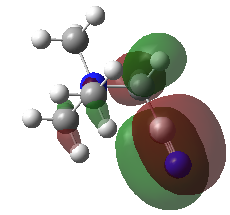 |
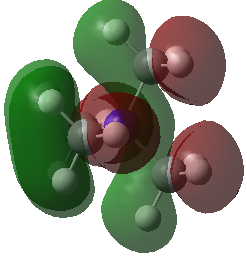
|
| LUMO | 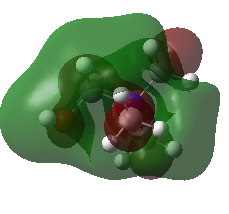 |
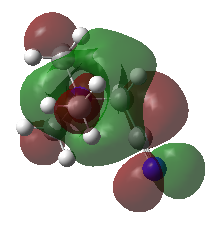 |
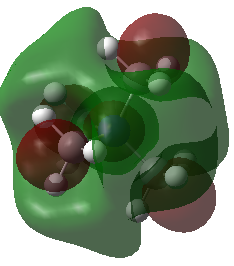
|
Note: The HOMO of [N(CH3)4]+ is triply degenerate.
| [N(CH3)3(CH2OH)]+ | [N(CH3)3(CH2CN)]+ | [N(CH3)4]+ | |
|---|---|---|---|
| HOMO | -0.48763 | -0.50047 | -0.57934 (degenerate) |
| LUMO | -0.12459 | -0.18183 | -0.13303 |
| HOMO-LUMO gap (KJ•mol-1) | 953 | 837 | 1172 |
The HOMO of [N(CH3)4]+ molecule is triply degenerate due to the Td symmetry. When one methyl group was replaced by -OH or -CN group, this degeneracy was broken and the structure became more stable due to the electronegative of O and N atom. The HOMO-LUMO gap also decreased.
Interesting aspect of [N(CH3)3(CH2OH)]+
During the optimization of [N(CH3)3(CH2OH)]+, there are two possible structure (Symmetry) for which O-H bond could be both periplanar or antiperiplanar to the next C-N bond. The result shows that periplanar structure are preferred since the two lone pair electrons in O atom is favoring on opposite site with two adjacent C-H bond. These could be explained by both steric effect and stabilization of C-H anti-bonding orbital.