Rep:Mod:AlexanderGrayCont
Organic Computaional Labs Continued, Part1
The Second Reaction Intermediate of a Glycosidation Reaction
As stated previously the results for the MM2 and MOPAC calculations of each isomer of the second reaction intermediate are shown below (These calculations were run on a new version of the software):
Pentahelicene |
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.1410 Bend: 15.2130 Stretch-Bend: 0.7546 Torsion: 9.4948 Non-1,4 VDW: -3.7163 1,4 VDW: 17.7977 Charge/Dipole: -4.6530 Dipole/Dipole: -0.7804 Total Energy: 36.2514 kcal/mol Calculation ended -------------------------------------------
Mopac Job: AUX RM1 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09967 (< 0.10000) Heat of Formation = -76.27765 Kcal/Mol -----------------------------------------
Pentahelicene |
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.0053 Bend: 13.4884 Stretch-Bend: 0.7191 Torsion: 9.9562 Non-1,4 VDW: -2.8698 1,4 VDW: 18.0192 Charge/Dipole: -7.7771 Dipole/Dipole: -2.4245 Total Energy: 31.1168 kcal/mol Calculation ended -------------------------------------------
Mopac Job: AUX RM1 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.08015 (< 0.10000) Heat of Formation = -79.45545 Kcal/Mol -----------------------------------------
The MOPAC calculations result for the Heat of Formation here is the same as the isomer from the other part, this shows that they are in fact the same molecule and that the bond does indeed exist in the previous part even though it is not visible.
Pentahelicene |
MM2 Calculation completed successfully ------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.6633 Bend: 19.5290 Stretch-Bend: 0.7749 Torsion: 9.1167 Non-1,4 VDW: -3.2021 1,4 VDW: 19.0162 Charge/Dipole: 3.3946 Dipole/Dipole: -1.5909 Total Energy: 49.7016 kcal/mol Calculation ended -------------------------------------------
Mopac Job: AUX AM1 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.08886 (< 0.10000) Heat of Formation = -62.63771 Kcal/Mol -----------------------------------------
Pentahelicene |
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.7503 Bend: 18.0616 Stretch-Bend: 0.8380 Torsion: 9.9783 Non-1,4 VDW: -2.6260 1,4 VDW: 19.5983 Charge/Dipole: 2.3600 Dipole/Dipole: -1.6307 Total Energy: 49.3298 kcal/mol Calculation ended -------------------------------------------
Mopac Job: AUX RM1 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09213 (< 0.10000) Heat of Formation = -54.08588 Kcal/Mol -----------------------------------------
For isomers 1,3 and 4 the additional bond adds strain to the molecule and so raises the total energy of the molecule and lowers the heat of formation.
Part 2, Module 1
Taxol
To test the NMR prediction capability's of the software a molecule of Taxol was generated, optimized and then run though an NMR analysis, this is shown below:
Pentahelicene |
The optimization log file can be found here: File:Log for new taxol.log
The NMR analysis for taxol using a Basis set of mpw1pw91/6-31G(d,p) can be found here DOI:10042/24302
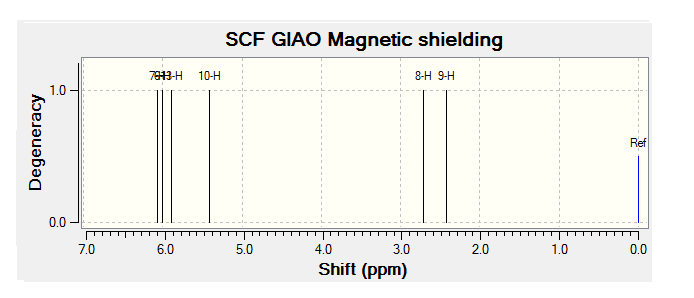
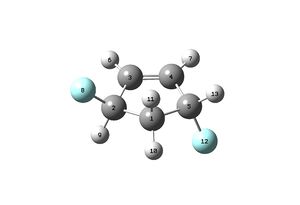
The H1 NMR spectrum for the Taxol Complex can be seen below:
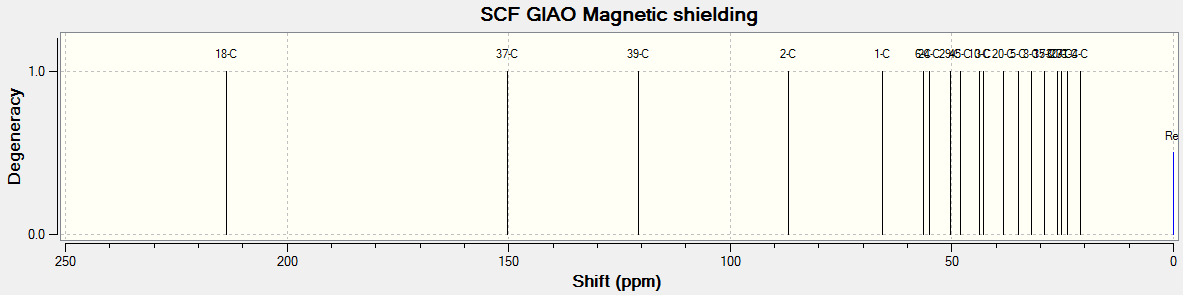
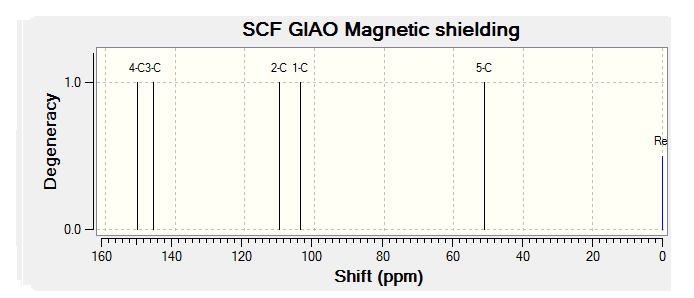
The C13 NMR spectrum for the Taxol complex can be seen below:
The literature[1] NMR data has been tabulated below with the corresponding calculated NMR data for comparison:
| Liturature NMR (ppm) | Calculated NMR (ppm) |
| 218.8 | 213.6 |
| 144.6 | 150.3 |
| 125.3 | 120.7 |
| 72.9 | 87.0 |
| 56.2 | 65.8 |
| 52.5 | 56.5 |
| 48.5 | 55.0 |
| 46.8 | 50.4 |
| 45.8 | 50.4 |
| 39.8 | 38.3 |
| 38.8 | 42.9 |
| 35.9 | 41.5 |
| 32.7 | 35.9 |
| 28.8 | 34.9 |
| 28.3 | 31.9 |
| 26.9 | 28.5 |
| 25.7 | 29.1 |
| 23.9 | 26.3 |
| 21.0 | 23.9 |
| 18.7 | 21.0 |
As can be seen from the table there is generally good agreement between the calculated and literature results for the C13 NMR spectrum. In general the calculated results are a few ppm higher than the literature values, this is most likely due to a to smaller basis set being used or the reference conditions being used are not exactly the same as the literature. As the basis set is improved then the accuracy of the computed NMR will increase and this should account for the difference.
Mini Project
For this mini project the molecule shown below was chosen from literature[2] so that its reported NMR could be checked to confirm that the different isomers had been assigned correctly. The first set was to construct the molecule with the different possible isomer and then generate predicted NMRs this is shown below:
 The reaction scheme for the formation of the products being looked at in this project
The reaction scheme for the formation of the products being looked at in this project
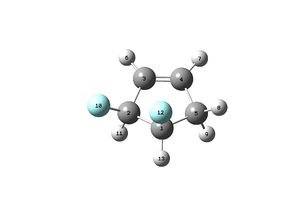
This is a Jmol of one of the isomers present:
Pentahelicene |
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 105: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.4999 Bend: 5.6308 Stretch-Bend: -0.1001 Torsion: 0.6953 Non-1,4 VDW: -0.2984 1,4 VDW: 1.7252 Dipole/Dipole: 3.6138 Total Energy: 11.7665 kcal/mol Calculation completed ------------------------------------
The same molecule was optimized on Gaussview the following log file was generated from this: File:OPT 1.LOG The optimized molecule was then further optimized using the 6-31G(d,p) basis set the file ca be found here: File:OPT 2.LOG
For each of the isomers the molecule was optimized using gaussian after being modified from the previously optimized molecule, the files from each optimization can be found with the corresponding DOI links.
Optimization of isomer 2 DOI:10042/24282
Optimization of isomer 3 DOI:10042/24284
Optimization of isomer 4 DOI:10042/24285
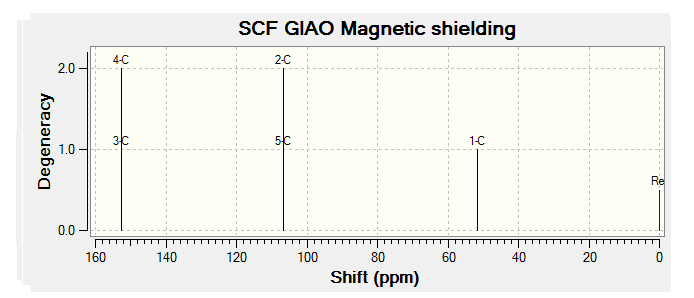
On these optimized molecules an NMR analysis was run using the B3LYP - 6-31G(d,p) basis set, the NMR spectra obtained are displayed below and the files for these calculations can be found at the following links:
NMR analysis for isomer 1 DOI:10042/24286
NMR analysis for isomer 2 DOI:10042/24287
NMR analysis for isomer 3 DOI:10042/24288
NMR analysis for isomer 4 DOI:10042/24289
A second NMR analysis was then run this time using a much larger basis set (cc-pVTZ) to investigate if the predicted NMR could be improved, the results have been tabulated below and the files for the calculations can be found here:
Larger basis set NMR analysis for isomer 1 DOI:10042/24290
Larger basis set NMR analysis for isomer 2 DOI:10042/24291
Larger basis set NMR analysis for isomer 3 DOI:10042/24292
Larger basis set NMR analysis for isomer 4 DOI:10042/24293
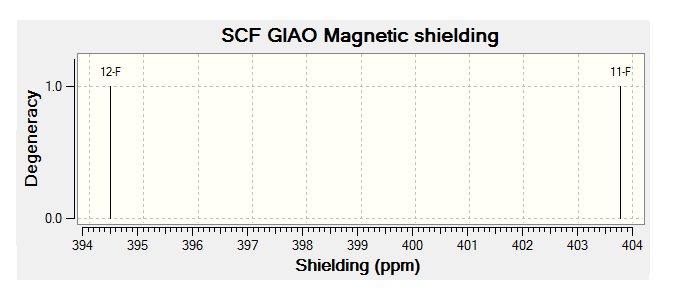
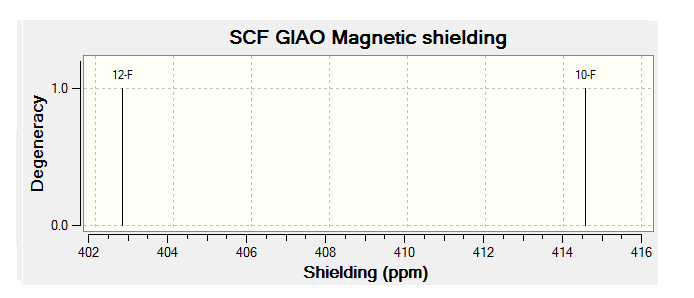
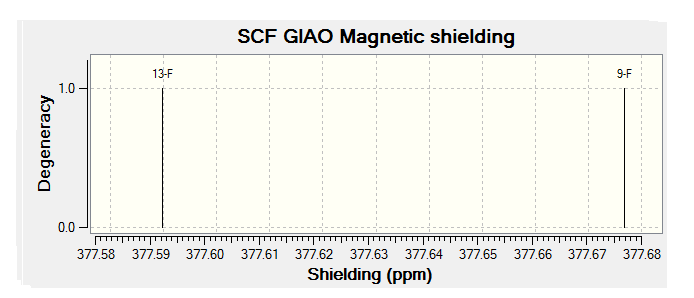
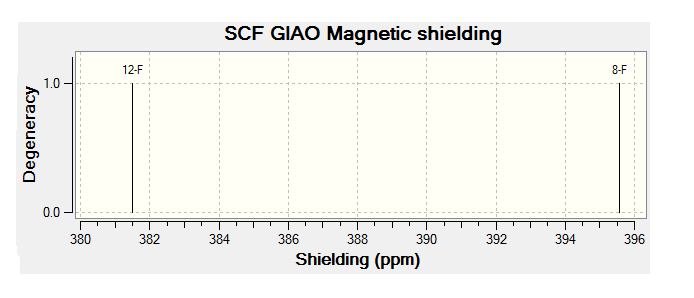
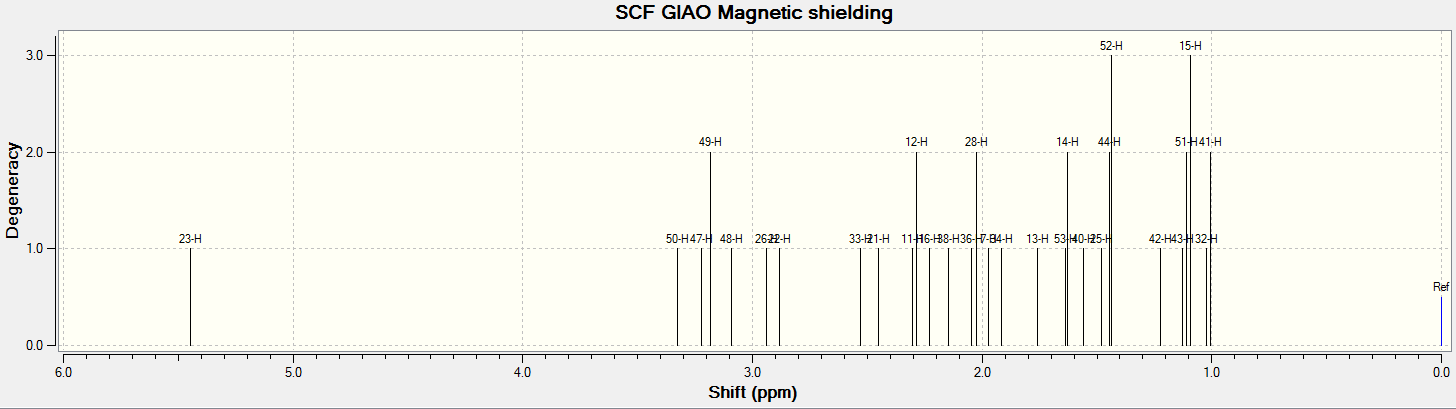
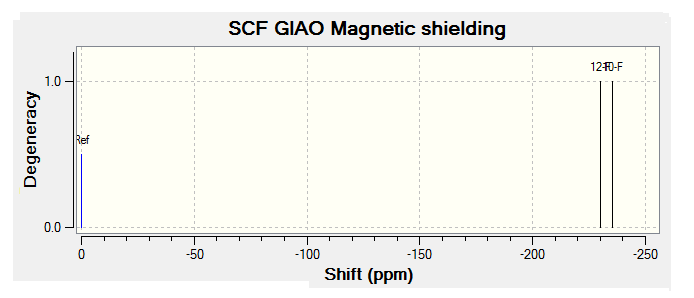
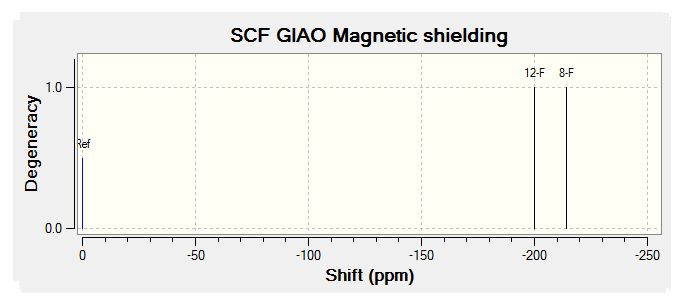
For all of the spectra in this table the relevant reference molecule has been used apart from in the case of the F19 NMR where there was no references in Gaussian.
The table below contains the same NMR spectra as above but they have been calculated using a higher basis set. The spectra here have been shown to correlate better to the recorded NMR found in literature. To allow analysis of the F19 NMR a reference molecule was made and the shielding constant (168.5547) was manually entered into Gaussian, the molecule was one of CFCl3 chosen as it is the same one used in the literature. The output files for the optimization and NMR analysis for CFCl3 can be found at their respective links. The ppm values obtained from Gaussian have been found to be slightly too low, this could be down to the reference calculated or some other error of Gaussian.
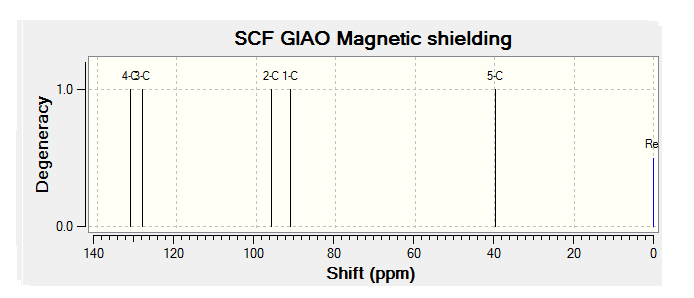
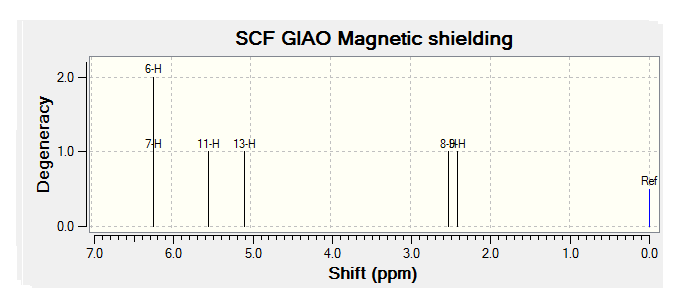
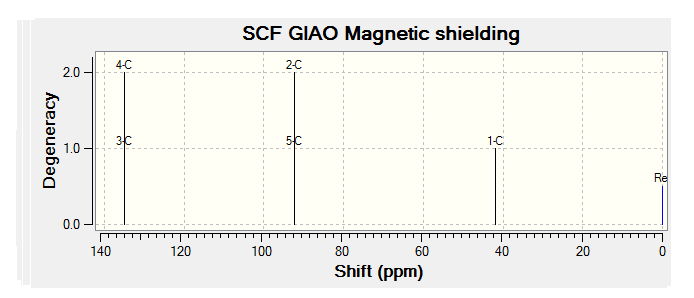
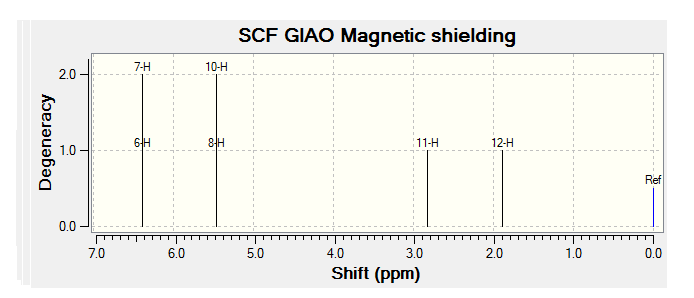
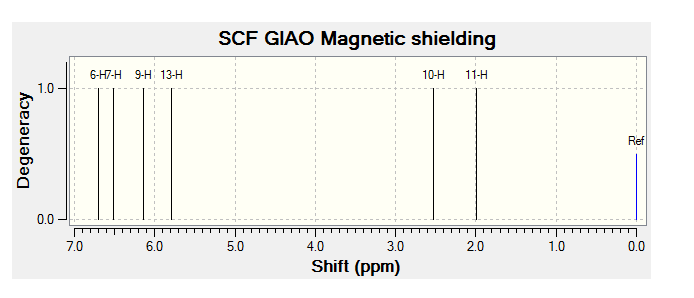
| JMol of Relevant Molecule | Flourene NMR Spectra | Carbon NMR Spectra | Hydrogen NMR Spectra | |||
|---|---|---|---|---|---|---|
|
 |
 |
| |||
|
 |
 |

| |||
|
 |
 |

| |||
|
 |
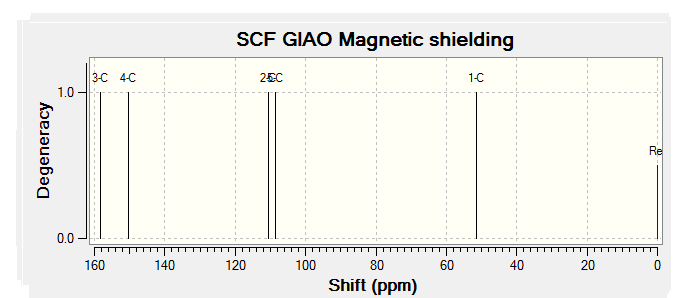 |
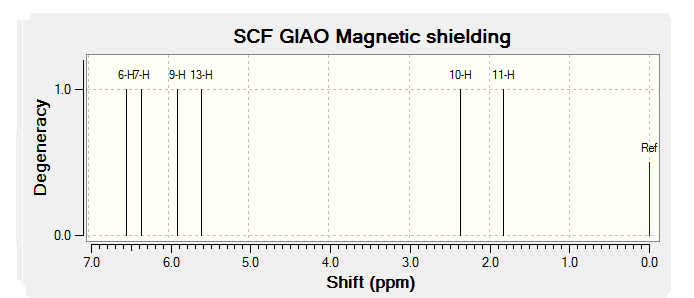
|
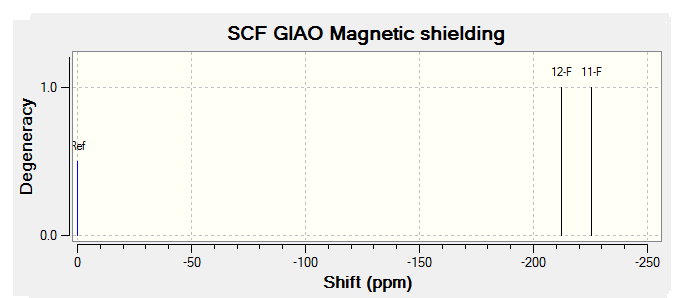
The NMR data from Literature[3] has been tabulated below:
| Isomer 1 (ppm) | Isomer 2 (ppm) | Isomer 3 (ppm) | Isomer 4 (ppm) |
|---|---|---|---|
| 2.00-3.30 (m, 2H); | 2.00-2.90 (m, 2H) | 2.06 (tdt, 1H) | 2.35 (tt, 2H) |
| 5.22 (ddd, 1H); | 5.68 (ddm, 1H) | 2.40-3.10 (m, 1H) | 5.78 (ddm, 2H) |
| 5.6 (ddm, 1H); | 6.00-6.80 (m, 3H) | 4.90-5.90 (m, 2H) | 5.80-6.40 (m, 2H) |
| 5.85 (dm, 1H); | 6.18-6.40 (m, 2H) | ||
| 6.80 (m, 1H); |
By comparing the chemical shift recorded to the predicted shifts above it can be seen that there is some agreement in the order and approximate number of chemical shifts present, taking into account that some peaks recorded as multiplets of a relative number of hydrogen's more than one correlate to multiple predicted peaks. For example if we look at isomer 1 it can be seen for the predicted NMR graph that there should be 6H's present, this is the case, and that there are expected to be 2 H's between 2-3ppm, again the case. The peaks seen in literature at 5.2, 5.6 and 5.8 have also been correctly predicted by the NMR prediction software, however the peak with chemical shift of 6.8 has been predicted at 6.2 ppm, a significantly lower value, this implies that the software has not correctly predicted the chemical shift of the hydrogen attached to an alkene and with a fluorine on a neighboring carbon atom. However the general good agreement shows that isomer one has been correctly predicted by the software and assigned correctly by the literature.
Unfortunately the other isomers do not show as close a similarity to the literature, it is stated that the purity of isomer 2 (compound 3 in literature) is not of complete purity, this can be seen from the wide ranges in ppm recorded and the number of multiplets. The predicted spectra shows 2 H's at roughly 2.5 ppm, this can be correlated to the literature values between 2.0-2.9ppm the rest of the spectra does not however correlate to the predicted spectra any more than any other isomer and so the NMR data, stated unreliable in the literature should not been be for further analysis.
The remaining two isomer can be seen to relate to the predicted NMR spectra but there is still a reasonably large margin of error as far as the chemical shifts are concerned but it can be said that the H1 NMR spectra of compounds have been assigned correctly.
| Isomer 1 (ppm) | Isomer 2 (ppm) | Isomer 3 (ppm) | Isomer 4 (ppm) |
|---|---|---|---|
| -176 (ddm, 1F); | -159 (m, 1F) | -161 (m) | -171 (m) |
| -187 (m, 1F); | -167 (m, 1F) |
The literature[4] values for the F19 NMR, seen above, can be seen to have a higher (less negative) chemical shift than the predicted NMR values despite the corrections made. looking at the distribution of the peaks however it is possible to see that the for isomer 1 and 2 both methods show 2 distinguishable peaks with the spacing between peaks being larger for isomer 1 than 2. For isomer 2 only 1 peak is expected as the molecule is symmetric and so both flourines are in the same environment, only one peak is seen in the actual NMR, this is a multiplet probably due to coupling with the hydrogen atoms. Isomer 4 has been predicted to show 2 peaks that should be differentiable, this however is not the case, the chemical shift of isomer 3 is predicted to be higher (less negative) than that of isomer 4, although the values are not the same the relationship is and so it can be said that the correct assignment has been made.
J coupling
To look at the spin coupling for each molecule the following data was extracted from the log files of each of the diflorinated molecules, the original log files for each can be found above. The data below shows the couplings between each atom, the labeling system relates the the molecule above the data, the light blue atoms are fluorine atoms, the white hydrogen and the grey carbon.
Unfortunately as the literature does not provide many coupling constants there is very little comparative analysis possible here. Values of interest have been tabulated below each set of data.
Isomer 1
Total nuclear spin-spin coupling J (Hz):
1 2 3 4 5
1 0.000000D+00
2 0.342866D+02 0.000000D+00
3 0.895875D+01 0.387721D+02 0.000000D+00
4 0.432962D+01 0.357276D+01 0.814371D+02 0.000000D+00
5 0.286622D+02 0.761418D+01 0.168366D+00 0.277392D+02 0.000000D+00
6 0.620195D+01 0.250870D+01 0.154230D+03 0.241025D+01 0.890794D+01
7 0.641252D+01 0.975844D+01 0.233016D+01 0.151755D+03 0.453581D+01
8 -0.356953D+01 0.490619D+01 0.420895D+01 -0.373378D+01 0.118209D+03
9 -0.562801D+01 -0.180511D+01 0.340892D+01 -0.614534D+01 0.112126D+03
10 0.131662D+03 -0.504874D+01 -0.296543D+00 0.105109D+01 -0.136180D+01
11 -0.228247D+03 0.199291D+02 0.648570D+01 0.435993D+01 0.161753D+02
12 0.158580D+02 -0.221931D+03 0.181425D+02 0.275804D+01 0.247006D+01
13 -0.387550D+01 0.133542D+03 -0.281012D+01 0.386991D+01 -0.136352D+01
6 7 8 9 10
6 0.000000D+00
7 0.570798D+01 0.000000D+00
8 -0.239625D+01 0.333843D+01 0.000000D+00
9 -0.377890D+01 0.273722D+01 -0.164174D+02 0.000000D+00
10 0.152419D+00 -0.118905D+00 0.752471D+01 0.513857D+01 0.000000D+00
11 0.114526D+01 0.180493D+01 0.496252D+00 0.209070D+02 0.485841D+02
12 0.243819D+01 -0.284005D+01 0.609967D+00 0.104561D+02 0.227008D+02
13 0.225434D+01 -0.251199D+01 0.230756D+01 0.300313D+01 0.361997D+01
11 12 13
11 0.000000D+00
12 -0.656033D+01 0.000000D+00
13 0.173949D+02 0.493934D+02 0.000000D+00
From this data the most significant couplings have been tabulated below:
| Coupling atoms / source | Coupling (Hz) |
|---|---|
| F-F coupling / Computed | 22 |
| F-F coupling / Literature | 18 |
The values obtained here are similar if not exactly the same as literature, considering that in the NMR analysis the chemical shifts where slightly larger than there reported counterparts this difference would be predicted.
The same file for isomer 2 is below:
Total nuclear spin-spin coupling J (Hz):
1 2 3 4 5
1 0.000000D+00
2 0.298057D+02 0.000000D+00
3 0.360005D+01 0.383120D+02 0.000000D+00
4 0.140841D+01 0.568185D+01 0.847547D+02 0.000000D+00
5 0.272228D+02 0.661319D+01 0.146319D+01 0.276719D+02 0.000000D+00
6 0.531289D+01 0.294077D+01 0.154554D+03 0.326074D+01 0.956471D+01
7 0.552333D+01 0.112712D+02 0.331786D+01 0.151972D+03 0.495994D+01
8 -0.540504D+01 0.397276D+01 0.386070D+01 -0.396731D+01 0.116736D+03
9 0.381514D+01 -0.113783D+01 0.287703D+01 -0.488835D+01 0.111760D+03
10 0.884900D+01 -0.227663D+03 0.174540D+02 0.474589D+01 -0.173807D+01
11 0.548960D+01 0.132178D+03 -0.215125D+01 0.257544D+01 -0.786536D+00
12 -0.224697D+03 0.112122D+02 -0.314235D+00 -0.149288D+01 0.150435D+02
13 0.145676D+03 0.312450D+00 0.540799D+01 0.585986D+01 0.177718D+01
6 7 8 9 10
6 0.000000D+00
7 0.541089D+01 0.000000D+00
8 -0.230815D+01 0.333160D+01 0.000000D+00
9 -0.411266D+01 0.255795D+01 -0.176851D+02 0.000000D+00
10 0.241296D+01 -0.149280D+01 0.181811D+00 0.359811D+01 0.000000D+00
11 0.215383D+01 -0.323173D+01 0.133489D+01 0.274682D+01 0.442501D+02
12 -0.119884D-01 -0.683563D-02 0.213811D+02 0.256981D+02 -0.570540D+01
13 0.288938D+00 0.320102D+00 0.497912D-01 0.396887D+01 -0.132042D+01
11 12 13
11 0.000000D+00
12 0.122457D+02 0.000000D+00
13 0.410809D+01 0.492388D+02 0.000000D+00
| Coupling atoms / source | Coupling (Hz) |
|---|---|
| F-F coupling / Computed | 5.7 |
| F-F coupling / Literature | None reported |
For isomer 3:
Total nuclear spin-spin coupling J (Hz):
1 2 3 4 5
1 0.000000D+00
2 0.273345D+02 0.000000D+00
3 0.351869D+01 0.357805D+02 0.000000D+00
4 0.351678D+01 0.356759D+01 0.851369D+02 0.000000D+00
5 0.273508D+02 0.784544D+01 0.356356D+01 0.357942D+02 0.000000D+00
6 0.415912D+01 0.232123D+01 0.153275D+03 0.317256D+01 0.115299D+02
7 0.415929D+01 0.115328D+02 0.317046D+01 0.153272D+03 0.232109D+01
8 0.126697D+01 0.766162D+00 0.282143D+01 -0.214675D+01 0.135968D+03
9 0.110154D+02 -0.221170D+03 0.175499D+02 0.268770D+01 -0.224820D+01
10 0.126219D+01 0.135964D+03 -0.214746D+01 0.281796D+01 0.765104D+00
11 0.120768D+03 -0.126794D+01 0.546578D+01 0.546538D+01 -0.126249D+01
12 0.108735D+03 -0.707120D+01 -0.156287D+00 -0.156513D+00 -0.707035D+01
13 0.110113D+02 -0.226057D+01 0.268852D+01 0.175542D+02 -0.221062D+03
6 7 8 9 10
6 0.000000D+00
7 0.509273D+01 0.000000D+00
8 -0.209240D+01 0.221891D+01 0.000000D+00
9 0.250491D+01 -0.292515D+01 0.110507D+02 0.000000D+00
10 0.221853D+01 -0.209470D+01 0.188537D+01 0.483661D+02 0.000000D+00
11 -0.246804D+00 -0.246888D+00 0.628814D+01 0.404899D+00 0.628769D+01
12 -0.122410D+00 -0.120443D+00 0.486518D+01 0.246650D+02 0.486525D+01
13 -0.293299D+01 0.250911D+01 0.483593D+02 0.121202D+02 0.110678D+02
11 12 13
11 0.000000D+00
12 -0.122699D+02 0.000000D+00
13 0.410527D+00 0.246816D+02 0.000000D+00
| Coupling atoms / source | Coupling (Hz) |
|---|---|
| F-F coupling / Computed | 12.1 |
| F-F coupling / Literature | None reported |
Isomer 4:
Total nuclear spin-spin coupling J (Hz):
1 2 3 4 5
1 0.000000D+00
2 0.291101D+02 0.000000D+00
3 0.376865D+01 0.360903D+02 0.000000D+00
4 0.327801D+01 0.501414D+01 0.846316D+02 0.000000D+00
5 0.293380D+02 0.528212D+01 0.261876D+01 0.337271D+02 0.000000D+00
6 0.452981D+01 0.290011D+01 0.152690D+03 0.334557D+01 0.114129D+02
7 0.428822D+01 0.108793D+02 0.372204D+01 0.153433D+03 0.387792D+01
8 0.126708D+02 -0.215381D+03 0.149397D+02 0.209481D+01 0.934862D+01
9 -0.600859D-01 0.134584D+03 -0.146461D+01 0.311426D+01 -0.920073D+00
10 0.117780D+03 -0.161334D+01 0.628257D+01 0.319068D+01 -0.485410D+01
11 0.109076D+03 -0.669288D+01 -0.176798D+00 0.856155D+00 -0.822490D+00
12 0.180918D+02 -0.366872D+01 0.296262D+01 0.774594D+01 -0.211300D+03
13 -0.195171D+00 0.546568D+01 0.457133D+01 -0.927065D+00 0.141349D+03
6 7 8 9 10
6 0.000000D+00
7 0.515899D+01 0.000000D+00
8 0.306287D+01 -0.256222D+01 0.000000D+00
9 0.222974D+01 -0.258174D+01 0.477343D+02 0.000000D+00
10 -0.172590D+00 0.135473D+00 0.311884D+00 0.566856D+01 0.000000D+00
11 -0.605838D+00 -0.518467D+00 0.236849D+02 0.415039D+01 -0.140235D+02
12 -0.434430D+01 0.255227D+01 0.190874D+02 0.799236D+01 0.176430D+02
13 -0.876659D+00 0.260831D+01 0.140782D+00 0.175197D+01 0.551757D-01
11 12 13
11 0.000000D+00
12 0.246213D+02 0.000000D+00
13 0.565082D+01 0.497731D+02 0.000000D+00
| Coupling atoms / source | Coupling (Hz) |
|---|---|
| F-F coupling / Computed | 19.1 |
| F-F coupling / Literature | None reported |
Vibrational Analysis
The optimized molecules for above where then run through vibrational analysis to produce IR spectra, the literature did not run IR spectra which these can be compared to, but it is a helpful technique to check that the compound is has been optimized and to compare any differences that arise (the IR spectra should contain the same peaks).
| Isomer Number | Calculated IR Spectrum |
|---|---|
| Isomer 1 | 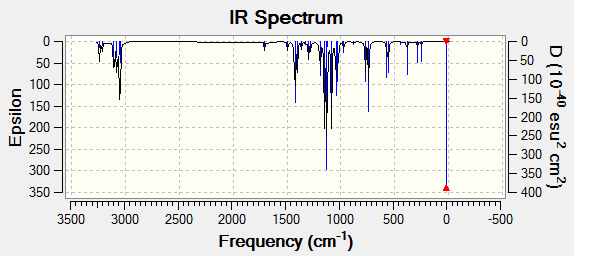
|
| Isomer 2 | 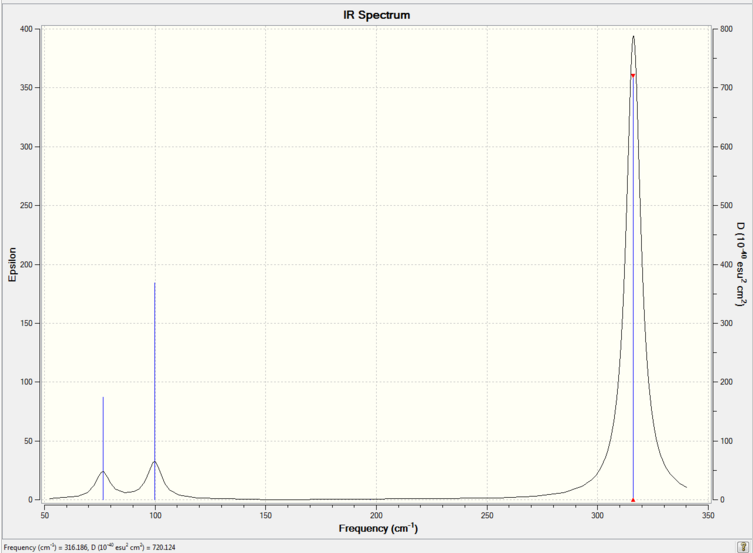
|
| Isomer 3 | 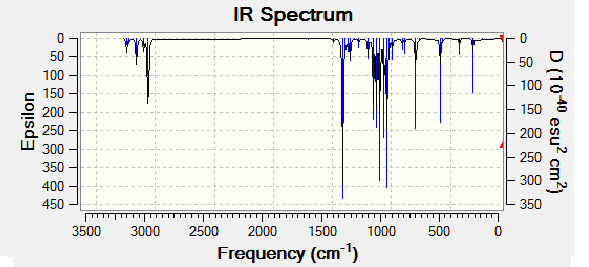
|
| Isomer 4 | 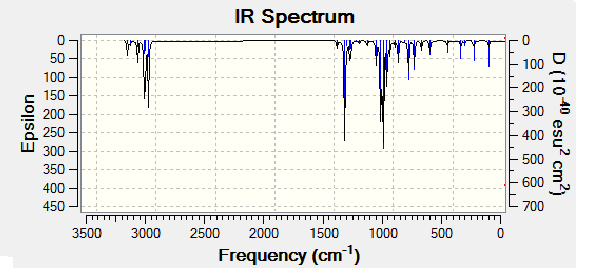
|
Comparison of the spectra shows that they are not all the same, however it is mainly the intensity of the peaks rather that there existence that changes. All the the molecules contain the expected peaks, Isomer 3 has additional peaks and much larger peaks compared to the other isomers, this is due to its high level of symmetry.
Conclusions
The data provided by the literature source has been proven to be of a poor quality and that computational analysis has provided more detailed evidence for the identification of each of the isomers produced. The literature did however get the assignment correct and so has been supported not disproved by this analysis.
References
- ↑ Wikipedia. [3.3] Sigmatropy within 1-vinyl-2-alkenyl-7,7-dimethyl-exo-norbornan-2-ols. The first atropselective oxyanionic Cope rearrangement, Normal 0 false false false EN-GB X-NONE X-NONE Leo A. Paquette , Neil A. Pegg , Dana Toops , George D. Maynard , Robin D. Rogers". http://pubs.acs.org/doi/abs/10.1021/ja00157a043
- ↑ Wikipedia. The fluorination of cyclopentadiene and 3,4-epoxycyclopentene, Dale F. Shellhamera, Michael Chua Chiacoa, Kelly M. Gallegoa, William S.C. Lowa, Barbara Cartera, Victor L. Heasleya, Robert D. Chapmanb, ". http://www.sciencedirect.com/science/article/pii/002211399403190B#
- ↑ Wikipedia. The fluorination of cyclopentadiene and 3,4-epoxycyclopentene, Dale F. Shellhamera, Michael Chua Chiacoa, Kelly M. Gallegoa, William S.C. Lowa, Barbara Cartera, Victor L. Heasleya, Robert D. Chapmanb, ". http://www.sciencedirect.com/science/article/pii/002211399403190B#
- ↑ Wikipedia. The fluorination of cyclopentadiene and 3,4-epoxycyclopentene, Dale F. Shellhamera, Michael Chua Chiacoa, Kelly M. Gallegoa, William S.C. Lowa, Barbara Cartera, Victor L. Heasleya, Robert D. Chapmanb, ". http://www.sciencedirect.com/science/article/pii/002211399403190B#