Mod:avogadro
See also: 1C comp-lab startup,1C Timetable,Laptop use,Programs,Module 1C Script,Writing up, Don't panic.
Avogadro
This is a general purpose 3D molecular modelling program, available as open source as version 1.1.[1] Bugs commonly encountered by students are documented here
Using the program
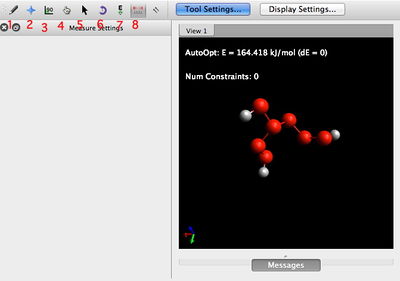
The main menu functions as follows. The most important tools are in bold.
- Ensure that the button labelled Tool Settings is toggled blue. This ensures that the tool settings appear on the left
- The draw tool. Use this to draw new atoms (carbon by default), dragging out a bond from an existing atom and releasing the mouse to drop a new atom.
- To close a ring, drag to the ring-closing atom and release (the hydrogens at the closed bond will be automatically removed).
- To delete atoms, click on them whilst pressing the right-mouse-button.
- To move (drag) atoms, place the mouse pointer on them, and drag to a new destination.
- For charged atoms (i.e. R-O(+)=C ), just specify the bond order and ignore the charge.
- Bonds can be created between two existing but non bonded atoms by specifying a bond order (single by default) and dragging the mouse between them.
- A right-mouse-click on a bond will remove it.
- The navigation tool, used to rotate and manipulate the structure
- Bond centric manipulation tool. Easier used than described!
- Manipulate (move) atoms. Use this to correct stereochemistry, alkene configuration, etc.
- Selection tool.
- Autorotation tool. Rotates the molecule according to a vector drawn with the mouse cursor.
- Energy optimisation tool. Use to calculate a molecular mechanics energy.
- use the following settings: Force Field=MMFF94s, Algorithm=Conjugate gradients
- Toggle 'Display Settings' on (the button will be blue) and tick Force in the display types. This shows the forces on each atoms during optimisation, which is complete when all the forces have vanished.
- Click start to obtain an energy. The optimiser remains in effect until you toggle to stop in order to edit the molecule, using e.g. move atoms (otherwise the auto-optimisation will rubber-band the atom and immediately return it to its start position).
- Measurement tool. Use to calculate lengths, angles and dihedral values.
Strategy
- The easiest way to create a molecule is to sketch it first in ChemDraw, save this as a .cdxml chemdraw file (do NOT use the older .cdx format, which crashes Avogadro), and then either drag-n-drop this file into the Avogadro viewing window, or simply open from within Avogadro, where it will be converted to an ~3D coordinate model of the molecule. You may have to correct stereochemistry by dragging atoms into the correct stereochemical position.
- To use Avogadro to start other types of calculation, use the Extensions menu item to create a job file for other programs (e.g Gaussian).
- Avogadro can also be used to read the outputs of other programs (e.g. Gaussian) and to display computed properties (vibrations, orbitals, etc).
Sources of data for Avogadro
Avogadro can accept data from the following sources:
- From 2D sketch files created as the .cdxml (Chemdraw) format.
- From 3D coordinate files created by other programs in a wide variety of formats.
- CIF format files generated by searches of the Cambridge crystallographic data base or by measurement of X-ray diffraction.
- From a URL (use eg http://www.ch.ic.ac.uk/rzepa/talks/76-79/ETP/SS-7.mol as an example)
- By specifying a chemical name (use e.g. aspirin as an example) or a protein name (use e.g. 1prc as an example)
Other uses of Avogadro
Avogadro is used in conjunction with a 2D sketching program ChemDoodle. These two together are used by the following courses:
Avogadro may be used to render and explore the MOs of a Gaussian optimised .fchk file. The orbitals display loads automatically when opening such a file and normally begins the user with 3/4 orbitals above and below the HOMO-LUMO region. Any other orbitals the molecule contains can be rendered and the quality changed via the menu in the bottom right hand corner.
Getting the program
Get the program [here] if you want to install it on your own computer.
References and citations
Please explore and report any strange behaviour to me. --Rzepa 09:29, 14 November 2012 (UTC)
See also: 1C comp-lab startup,1C Timetable,Laptop use,Programs,Module 1C Script,Writing up, Don't panic.
