Cxq2019
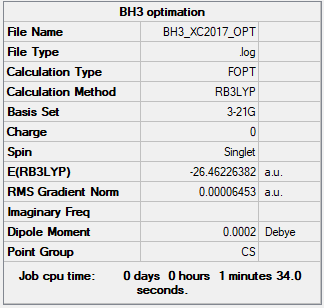
BH3 molecules
Use basic set: 3-21G
| Molecule name | BH3 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| E(RB3LYP) in a.u | -26.46226382 |
| Point Group | CS |
| B-H bond length/Å | 1.195 |
| H-B-H bond angle | 120.00 |
Item Value Threshold Converged?
Maximum Force 0.000168 0.000450 YES
RMS Force 0.000077 0.000300 YES
Maximum Displacement 0.000718 0.001800 YES
RMS Displacement 0.000336 0.001200 YES
Predicted change in Energy=-9.109419D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1945 -DE/DX = -0.0001 !
! R2 R(1,3) 1.1944 -DE/DX = 0.0 !
! R3 R(1,4) 1.1947 -DE/DX = -0.0002 !
! A1 A(2,1,3) 119.9989 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0151 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.986 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The optimization file is lined to here
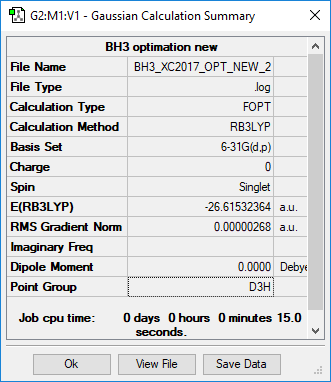
Use basic set: 6-31G(d,p)
| Molecule name | BH3 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -26.61532364 |
| Point Group | D3H |
| B-H bond length/Å | 1.192 |
| H-B-H bond angle | 120.00° |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-1.708777D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The optimization file is lined to here
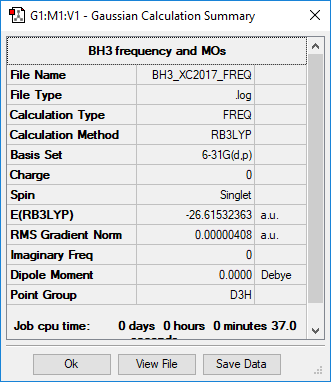
Frequency analysis
Ng611 (talk) 11:08, 15 May 2019 (BST) Good calculation. Remember that we just need to see the results from your frequency analysis.
Low frequencies --- -1.7446 -1.0515 -0.0054 2.8358 10.4988 10.5513
Low frequencies --- 1162.9876 1213.1768 1213.1795
Diagonal vibrational polarizability:
0.7180945 0.7179944 1.8414304
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2" E' E'
Frequencies -- 1162.9876 1213.1768 1213.1795
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5491 14.0547 14.0583
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
The optimization file is lined to here
Joml picture
BH3 |
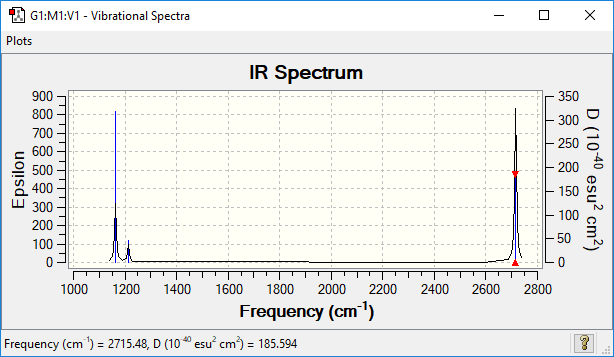
IR analysis
| wavenumber (cm-1 | Intensity (arbitrary units) | symmetry | IR active? | type |
| 1163 | 93 | A1 | yes | out-of-plane bend |
| 1213 | 14 | E | very slight | bend |
| 1213 | 14 | E | very slight | bend |
| 2582 | 0 | A1 | no | symmetric stretch |
| 2716 | 126 | E | yes | asymmetric stretch |
| 2716 | 126 | E | yes | asymmetric stretch |
Q: why are there less than six peaks in the spectrum, when there are obviously six vibrations?
A: The peaks shown in the IR spectrum shows that there is a change in dipole moment during the vibration. For the symmetric stretching vibration, there is no change in dipole moment. thus, it would not be shown in the spectrum. For those peaks, there is only a small change in dipole moment during the vibration.
Ng611 (talk) 11:09, 15 May 2019 (BST) That explains one peak, what about the others?
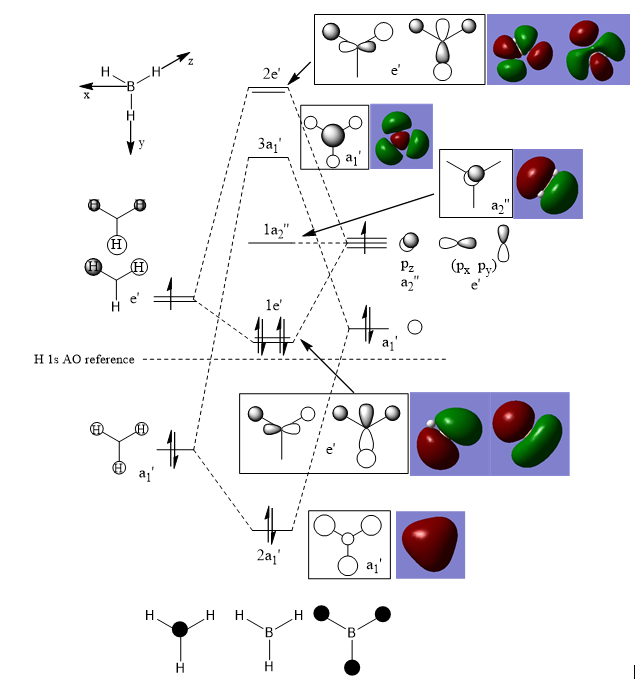
MOs for BH3 molecules
Q: Are there any significant differences between the real and LCAO MOs? What does this say about the accuracy and usefulness of qualitative MO theory?
A: For the real MO diagrams, orbitals sum up or cancel out with each other, the shape of the orbitals are not simply the sum of shapes in the literature, but it can still be recognised.
Ng611 (talk) 11:10, 15 May 2019 (BST) This is true, but how significant is this difference really?
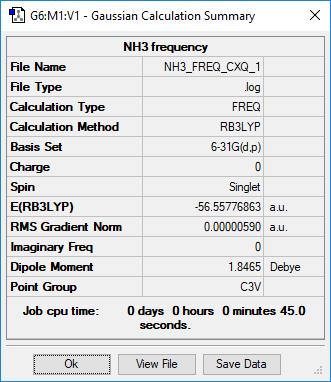
NH3 molecules
Low frequencies --- -8.5223 -8.4750 -0.0037 0.0335 0.1918 26.4067
Low frequencies --- 1089.7616 1694.1862 1694.1866
Diagonal vibrational polarizability:
0.1276540 0.1276552 3.2977130
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A1 E E
Frequencies -- 1089.7616 1694.1862 1694.1866
Red. masses -- 1.1800 1.0644 1.0644
Frc consts -- 0.8256 1.8001 1.8001
IR Inten -- 145.4217 13.5548 13.5550
Atom AN X Y Z X Y Z X Y Z
1 7 0.00 0.00 0.12 -0.07 0.00 0.00 0.00 0.07 0.00
2 1 0.00 -0.21 -0.53 0.76 0.00 0.00 0.00 0.15 0.26
3 1 0.18 0.11 -0.53 0.08 -0.39 0.22 0.39 -0.53 -0.13
4 1 -0.18 0.11 -0.53 0.08 0.39 -0.22 -0.39 -0.53 -0.13
The optimization file is lined to here
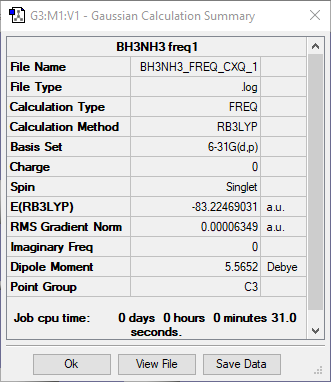
BH3NH3 molecules
Ng611 (talk) 11:11, 15 May 2019 (BST) You need to include ALL items (summary, low frequencies, log file, convergence table, jmol) for EVERY calculation.
Low frequencies --- -0.0017 -0.0015 -0.0008 17.5817 27.9112 40.2360
Low frequencies --- 266.5197 632.3594 639.4724
Diagonal vibrational polarizability:
2.5462260 2.5490111 5.0151955
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 266.5169 632.3591 639.4723
Red. masses -- 1.0078 4.9945 1.0451
Frc consts -- 0.0422 1.1767 0.2518
IR Inten -- 0.0000 13.9822 3.5411
Atom AN X Y Z X Y Z X Y Z
1 1 -0.36 0.02 0.00 0.00 0.04 0.29 0.14 0.03 -0.20
2 1 0.17 -0.32 0.01 -0.03 0.00 0.29 0.10 0.06 0.46
3 1 0.20 0.31 -0.01 0.03 -0.01 0.29 0.12 0.07 -0.26
4 1 -0.45 0.02 0.00 0.00 -0.01 -0.36 0.19 0.06 -0.26
5 1 0.24 0.38 -0.02 0.00 -0.02 -0.36 0.18 0.08 -0.33
6 1 0.20 -0.40 0.01 0.00 -0.02 -0.36 0.16 0.09 0.58
7 5 0.00 0.00 0.00 0.00 0.02 0.48 -0.03 -0.01 0.00
8 7 0.00 0.00 0.00 0.00 -0.01 -0.36 -0.04 -0.02 0.00
The optimization file is lined to here
Calculation for B-N bond energy
| E(BH3) in.a.u | -26.61532 |
|---|---|
| E(NH3) in a.u | -56.55777 |
| E(BH3NH3) in a.u | -83.22469 |
ΔE=E(BH3NH3)-[E(NH3)+E(BH3)] = -0.0516 a.u = -135 kJ/mol
Qː Based on the energy calculation is the B-N dative bond weak, medium or strong? What comparison have you made to come to this conclusion?
Aː the bond energy of B-N bond is 135 kJ/mol, which is a weak bond comparing to C-C bond (346 kJ/mol), B-B bond (293 kJ/mol), B-O bond (536 kJ/mol).
Ng611 (talk) 11:13, 15 May 2019 (BST) Where did you get your bond energies? You need to provide a citation (preferably to a literature source) to get the mark.
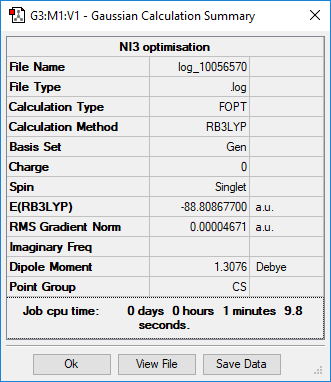
NI3 molecules
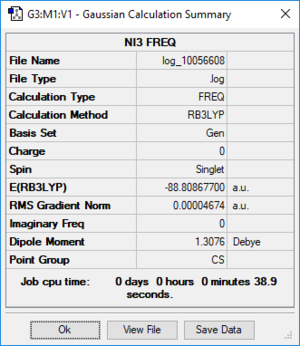
Item Value Threshold Converged?
Maximum Force 0.000061 0.000450 YES
RMS Force 0.000037 0.000300 YES
Maximum Displacement 0.000459 0.001800 YES
RMS Displacement 0.000285 0.001200 YES
Predicted change in Energy=-3.108653D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.184 -DE/DX = 0.0 !
! R2 R(1,3) 2.184 -DE/DX = 0.0 !
! R3 R(1,4) 2.184 -DE/DX = 0.0 !
! A1 A(2,1,3) 110.9144 -DE/DX = 0.0 !
! A2 A(2,1,4) 110.8498 -DE/DX = -0.0001 !
! A3 A(3,1,4) 110.9144 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 123.6745 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The optimisation file is lined to here
N-I bond lengthː 2.184 Å
NI3 |
Low frequencies --- -19.8898 -19.6985 -17.8980 -0.0005 -0.0004 -0.0001
Low frequencies --- 101.7538 102.1821 154.5874
Diagonal vibrational polarizability:
3.9894533 7.0946637 9.3153185
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A' A" A'
Frequencies -- 101.7534 102.1765 154.5866
Red. masses -- 110.8405 111.9304 98.9616
Frc consts -- 0.6762 0.6885 1.3934
IR Inten -- 0.6204 0.6941 1.3560
Atom AN X Y Z X Y Z X Y Z
1 7 0.20 -0.32 0.00 0.00 0.00 -0.36 -0.42 -0.27 0.00
2 53 0.28 -0.44 0.00 0.00 0.00 0.55 -0.26 0.43 0.00
3 53 -0.15 0.24 0.46 -0.26 0.39 -0.26 0.15 -0.20 0.44
4 53 -0.15 0.24 -0.46 0.26 -0.39 -0.26 0.15 -0.20 -0.44
The frequency file is lined to here
Projectː Ionic liquid
[N(CH3)4]+
Item Value Threshold Converged? Maximum Force 0.000010 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000143 0.001800 YES RMS Displacement 0.000055 0.001200 YES Predicted change in Energy=-7.767156D-09 Optimization completed. -- Stationary point found.
The optimisation file is lined to here
N(CH3)4 |
Low frequencies --- -0.0016 -0.0014 -0.0013 34.5756 34.5756 34.5756
Low frequencies --- 216.7144 316.0577 316.0577
Diagonal vibrational polarizability:
1.3864664 1.3864664 1.3864664
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2 T1 T1
Frequencies -- 216.7144 315.9830 315.9830
Red. masses -- 1.0078 1.0333 1.0333
Frc consts -- 0.0279 0.0608 0.0608
IR Inten -- 0.0000 0.0000 0.0000
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 0.00 -0.02 0.02 0.00 -0.01 0.01 0.00
2 1 0.20 0.00 -0.20 -0.22 0.03 0.21 0.28 0.02 -0.28
3 1 -0.20 0.20 0.00 0.18 -0.18 0.00 -0.30 0.30 -0.01
4 1 0.00 -0.20 0.20 -0.03 0.22 -0.21 -0.02 -0.27 0.27
5 6 0.00 0.00 0.00 0.02 -0.02 0.00 -0.01 0.01 0.00
6 1 -0.20 0.00 -0.20 0.21 -0.03 0.21 0.28 0.02 0.28
7 1 0.20 -0.20 0.00 -0.18 0.18 0.00 -0.30 0.30 0.01
8 1 0.00 0.20 0.20 0.03 -0.21 -0.21 -0.02 -0.27 -0.27
9 6 0.00 0.00 0.00 -0.02 -0.02 0.00 0.01 -0.01 -0.02
10 1 0.00 -0.20 -0.20 -0.03 -0.22 -0.21 0.02 0.03 0.03
11 1 -0.20 0.00 0.20 -0.22 -0.03 0.21 0.07 -0.02 -0.08
12 1 0.20 0.20 0.00 0.18 0.18 0.00 -0.05 -0.06 -0.04
13 6 0.00 0.00 0.00 0.02 0.02 0.00 0.01 -0.01 0.02
14 1 0.00 0.20 -0.20 0.03 0.22 -0.21 0.02 0.03 -0.03
15 1 0.20 0.00 0.20 0.22 0.03 0.21 0.07 -0.02 0.08
16 1 -0.20 -0.20 0.00 -0.18 -0.18 0.00 -0.05 -0.06 0.04
17 7 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
The frequency file is lined to here
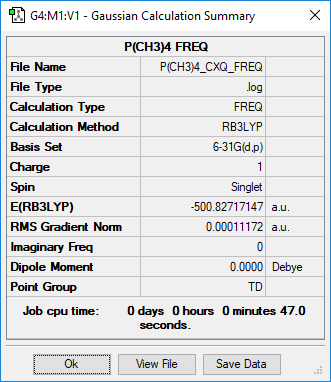
[P(CH3)4]+
Item Value Threshold Converged? Maximum Force 0.000122 0.000450 YES RMS Force 0.000033 0.000300 YES Maximum Displacement 0.000798 0.001800 YES RMS Displacement 0.000324 0.001200 YES Predicted change in Energy=-1.864576D-07 Optimization completed. -- Stationary point found.
The optimisation file is lined to here
P(CH3)4 |
Low frequencies --- 186.8775 211.6286 211.6286
Diagonal vibrational polarizability:
3.5426272 3.5426271 3.5426271
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2 T1 T1
Frequencies -- 186.8775 211.5665 211.5665
Red. masses -- 1.0078 1.0255 1.0255
Frc consts -- 0.0207 0.0270 0.0270
IR Inten -- 0.0000 0.0000 0.0000
Atom AN X Y Z X Y Z X Y Z
1 6 0.00 0.00 0.00 -0.02 0.01 0.00 -0.01 0.00 0.01
2 1 0.00 -0.20 0.20 -0.03 0.18 -0.17 -0.02 -0.25 0.25
3 1 0.20 0.00 -0.20 -0.18 0.03 0.18 0.22 0.00 -0.22
4 1 -0.20 0.20 0.00 0.15 -0.15 0.00 -0.24 0.24 0.03
5 6 0.00 0.00 0.00 0.01 -0.01 0.00 -0.02 0.00 -0.01
6 1 -0.20 0.00 -0.20 0.24 -0.03 0.24 0.15 0.00 0.15
7 1 0.20 -0.20 0.00 -0.22 0.22 0.00 -0.18 0.18 -0.03
8 1 0.00 0.20 0.20 0.02 -0.24 -0.24 -0.03 -0.17 -0.18
9 6 0.00 0.00 0.00 0.02 0.01 0.00 0.01 0.00 0.01
10 1 0.00 0.20 -0.20 0.03 0.17 -0.17 0.02 -0.24 0.24
11 1 0.20 0.00 0.20 0.18 0.03 0.17 -0.22 0.00 -0.22
12 1 -0.20 -0.20 0.00 -0.14 -0.14 0.00 0.24 0.24 0.03
13 6 0.00 0.00 0.00 -0.01 -0.01 0.00 0.02 0.00 -0.01
14 1 0.00 -0.20 -0.20 -0.02 -0.25 -0.25 0.03 -0.17 -0.17
15 1 -0.20 0.00 0.20 -0.24 -0.03 0.24 -0.14 0.00 0.14
16 1 0.20 0.20 0.00 0.22 0.22 0.00 0.18 0.17 -0.03
17 15 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00
The frequency file is lined to here
Charge distribution of [N(CH3)4]+ cation and [P(CH3)4]+ cation
| Ion name | [N(CH3)4]+ | [P(CH3)4]+ |
|---|---|---|
| Charge on central atom (N/P) | -0.295 | +1.666 |
| Charge on each C atom | -0.483 | -1.060 |
| Charge on each H atom | +0.269 | +0.298 |
| Element name | Electronegativity |
|---|---|
| N | 3.04 |
| P | 2.19 |
| C | 2.55 |
| H | 2.2 |
According to two tables above, it can be seen that the charge on H atom in both of the cation is similar. However, there is a huge difference in change on C atom and central atom. For the [N(CH3)4]+ ,the central atom is N and the charge on it is -0.295, while the central atom P in [P(CH3)4]+ has a positive charge (+1.666). This may due to their electronegativities shown in table above. The electronegativity for N is bigger than C while P has a smaller electronegativity than C. P is in the third row of the periodic table, it is larger and more polarisable than N which is in the 2nd row.
N atom in the [N(CH3)4]+ cation is less negative than the C atom in this atom. it may be because N is surrounded by 4 C atom which is slightly more electronegative than those H atoms surround by the C atom.
Ng611 (talk) 11:19, 15 May 2019 (BST) What about the effect of symmetry on charge distribution?
Validity of traditional description for [N(CH3)4]+ cation
In the validity of traditional description for [N(CH3)4]+ cation, there is a positive charge on N atom calculated by No. of valance electrons subtract the No. of non-bonding electrons and half of No. of bonding electrons.
However, according to the data shown in the previous section ,the positive charge actually sits on these H atoms.
This may be caused by the difference in ability to attract electrons for atoms (electronegativity). In the traditional description, the electrons are shared evenly between two atom which is quite different in real life. In the real life, the bonding electrons are attracted closer to the atom which has larger electronegativity.
Ng611 (talk) 11:20, 15 May 2019 (BST) Your answer doesn't make sense. How do you reconcile your formal electron counting picture (in your first para) with the 'electron sharing' picture (which requires diffuse, delocalised electrons) in your second para?
MOs for [N(CH3)4]+ cation
MO 7: Bonding character between central atom and ligand orbital, and anti-bonding character though space between ligands .
MO 10: Bonding character between central atom and the ligand orbital and ligand-ligand orbital though space.
Ng611 (talk) 11:22, 15 May 2019 (BST) Why use this FO? Why not the FO you used in MO7? That one would make more sense to be honest.
The MO 21 is the HOMO for this cation, and there is bonding character between ligand orbital but anti-bonding character though space between ligands.
Ng611 (talk) 11:24, 15 May 2019 (BST) In general, good LCAO analysis. Some more detail in your description of the orbital interactions would have improved your answer

![MO7 for the [N(CH3)4]+ cation.](/images/thumb/a/a7/MO7_N%28CH3%294_0510_1.PNG/500px-MO7_N%28CH3%294_0510_1.PNG)
![MO10 for the [N(CH3)4]+ cation.](/images/thumb/f/f5/MO10_N%28CH3%294_0510_1.PNG/500px-MO10_N%28CH3%294_0510_1.PNG)
![MO21 for the [N(CH3)4]+ cation.](/images/thumb/4/4b/MO21_N%28CH3%294_0510_1.PNG/500px-MO21_N%28CH3%294_0510_1.PNG)