AMBER
This method is specially designed for treating large biomolecules.
AMBER : a Molecular Mechanic's (MM) method
Principle of MM
This method is the simplest and least computationally demanding calculations, because it consists to apply some Newton's mechanics in our calculations. Indeed, the molecule is modelled as a framework of spherical atoms which are bound together with elastic bonds which are described as springs.
Different MM could be applied and they differ in how consider the energy terms, e.g. which approximations we can consider in a specific case. Indeed the power of MM lies in the fact that you can specify many parameters and functions in order to get the best way to simulate your molecule. Thus the parameters that you can type in and the choice of the MM method depends of the type of molecule you have.
Advantages and disadvantages
The electrons are neglected in this method so electronically excited states can not be treated without other specific parameters.
But the cost of these calculations is very low which makes it very attractive for calculations of large systems like proteins. And a good way to increase the accuracy of this method is to couple this one with the quantum mechanic.
Advice break
If you want More explanations about Molecular Mechanics in Gaussian click on the link and you will find some theory concerning MM with GAUSSIAN.
Moreover Molecular Mechanics Methods page on the GAUSSIAN website will give you a lot of information about running some calculations with MM and some errors that can appear.
Principle of AMBER
AMBER uses simple harmonic functions in order to modelise stretches and bends, cosine for torsion, and the standard functions for electrostatic and Van der Waals interactions.
The Amber force field as described in A 2nd Generation Force Field. The actual parameters (parm96.dat) have been updated slightly since the publication of this paper. We use this current version from the Amber web site.
How to run an AMBER calculation ?
General case
Molecular Mechanics calculations use atom types to determine the functions and parameters which make up the force field. For a single element, such as carbon, there can be many different MM atom types. Which one to use depends on aspects such as the hybridization, chemical environment, and so on.
Gaussian will attempt to assign atom types automatically for UFF and Dreiding calculations. However, Amber calculations require that all atom types be explicitly specified within the molecule specification section, as in these examples:
C-CT- sp3 aliphatic C C-CA- sp2 pure aromatic (benzene) H-HC- H aliphatic bonded to C without electron withdrawing group H-HA- H aromatic bonded to C without electron withdrawing group H-H1- H aliphatic bonded to C with 1 electron withdrawing group
Advice break
Consult the Amber paper for definitions of atom types and their associated keywords.
GaussView associates to each atom the atoms type automatically. This is particularly important because AMBER calculation corresponds to the association of a potential to each kind of bond length and angle (C CM H4 different to C CT H1 for example).
But all the different combinations of atoms’ type are not available directly in the software, so some MM parameters could be missing like, and so an error message appears in our output file :
Include all MM classes Bondstretch undefined between atoms 19 21 C-HA Angle bend undefined between atoms 15 19 21 CA-C-HA Angle bend undefined between atoms 16 19 21 CA-C-HA MM function not complete
It is possible to defined this different bond length and angles missing thanks to the Amber paper.
Thus, when you want to specify so such kind of relation between the atoms you need to insert the MM keyword amber=hardfirst in the route job and specify the different relations between the atoms at the end of the input file :
HrmStr1 C H1 367.0 1.080 so as to define a missing bond length HrmBnd1 CT C H1 50.0 120.0 so as to define a missing angle
Case of an ONIOM calculation
Run a calculation
As in the general case it is possible that such parameters (bond length, angles) were not already in the data base, ONIOM does not change this, but an other problem appears : how to define the frontier between the different layers ?
Indeed take the example of an ONIOM calculation with 2 different layers.
Two different calculations will concern the low level of theory : a calculation on the real system and a calculation on the model system.
If the there is no connection between the Model system and the surroundings there is no difference between the general case and this one, but if there are some connections between the model system and the real system new problems appear.
As usual with ONIOM, Gaussian replaces the different groups bonded to the high level part by an hydrogen, but in the case of an AMBER calculation Gaussian has to specify what kind of hydrogen it is. So as to define this kind of hydrogen GaussView associates an hydrogen which keeps the effect of the surroundings group attached to the model system, and so this generates new missing parameters involving this new hydrogen.
Let's take an example in order to explain :
Try to draw this molecule, and select the 2 layers as showed in the picture
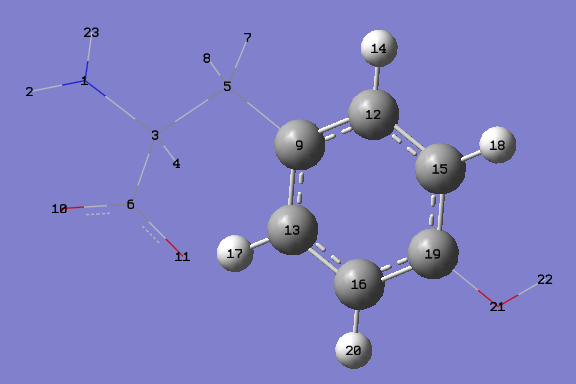
Now try to run a single point energy calculation with ONIOM : B3LYP with a 6-31g* basis set as a high level of theory and AMBER as a low level of theory
#p oniom(b3lyp/6-31g(d):amber) geom=connectivity
You should get an error message in your output file
Generating MM parameters for model system. Include all MM classes Bondstretch undefined between atoms 19 21 C-HA [H,L] Angle bend undefined between atoms 15 19 21 CA-C-HA [H,H,L] Angle bend undefined between atoms 16 19 21 CA-C-HA [H,H,L] MM function not complete
So some parameters are missing and they are all related to the new Hydrogen introduce at the frontier between high and low level. The reason of the absence of these parameters is that the Amber force field has been developed specifically for biochemical systems, in which these particular sequences of atom types do not occur.
First have a look at the different type of hydrogen that GaussView used so as to replace the surroundings groups
N-N3- 0 -1.2966 -1.02917 0. L H-H- 0 -0.29704 -1.05884 0. L C-CT- 0 -1.98355 0.24664 0. L H-H1- 0 -2.60838 0.32344 -0.88982 L C-CT- 0 -2.87017 0.39276 1.23214 L H-HA 9 0. 0. C-C- 0 -0.99307 1.40225 0. L H-HC- 0 -3.61441 -0.40356 1.24137 L H-HC- 0 -2.2574 0.32711 2.1312 L C-CA- 0 -3.56685 1.73193 1.19494 H O-O2- 0 0.21688 1.18672 0. L O-O2- 0 -1.39117 2.56499 0. L C-CA- 0 -4.4397 2.09581 2.22728 H C-CA- 0 -3.33991 2.60965 0.12813 H H-HA- 0 -4.61639 1.41244 3.05787 H C-CA- 0 -5.08563 3.33742 2.19279 H C-CA- 0 -3.98584 3.85126 0.09364 H H-HA- 0 -2.66033 2.32634 -0.67562 H H-HA- 0 -5.76521 3.62073 2.99653 H C-C- 0 -4.85869 4.21514 1.12597 H H-HA- 0 -3.80915 4.53463 -0.73696 H O-OH- 0 -5.48616 5.42128 1.09247 L H-HA 19 0. 0. H-HO- 0 -6.06002 5.56669 1.84821 L H-H- 0 -1.82207 -1.87998 0. L
As you can notice GaussView attributes the hydrogen HA in order to replace the carbon 5 and the group attached. As you can read in the Amber paper : HA corresponds to an H aromatic bonded to a carbon without electron withdrawing group.
The Oxygen 21 is also replace by an HA in the model system.
So these new sequences of atoms have to be defined at the end of the input file.
To specify new parameters, use the Amber=HardFirst keyword, and then add them to the input file in a separate section.
The new route job becomes :
#p oniom(b3lyp/6-31g(d):amber=hardfirst) geom=connectivity
And the input file ended with his new section :
HrmStr1 C HA 367.0 1.080 HrmBnd1 CA C HA 35.0 120.0
The data were found in the Amber paper.
Thus, with this addition the calculation is successful and the energy that I got with my geometry was :
ONIOM: calculating energy. ONIOM: gridpoint 1 method: low system: model energy: 0.006551530903 ONIOM: gridpoint 2 method: high system: model energy: -232.248031717645 ONIOM: gridpoint 3 method: low system: real energy: 1.163471769804 ONIOM: extrapolated energy = -231.091111478743
Optimisation
Opt=QuadMacro takes into account the coupling between atoms in the model system and the atoms only in the MM layer in order to produce more accurate steps than the regular microiterations algorithm.
Opt=Quadmac is mainly used for problem convergence cases, which does a quadratic step in the coordinates of all the atoms. Such an optimization can use either an updated approximate Hessian for the internal coordinates or an analytically computed Hessian (see the next bullet). It computes products of the low-level (MM) Hessian with vectors as needed.
Errors fair
Just a non exhaustive list of errors possible.
Type of atom missing
When you get this kind of error message at the end of your output file
AtZEff= 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 NQMom= 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 NMagM= 2.7928460 0.0000000 2.7928460 2.7928460 0.0000000 2.7928460 2.7928460 0.0000000 2.7928460 Generating MM parameters. Missing atomic parameters for atom 16 IAtTyp= 20000000 Missing atomic parameters. Error termination via Lnk1e
It corresponds to the fact that GaussView did not assign a type of atom to the atom 16 of your system. GaussView does not recognize the environment of the atom 16 and so was not able to enter the type of atom.
0 2 C-CA- -1.72939065 0.87813619 0.00000000 C-CA- -0.33423065 0.87813619 0.00000000 C-CA- 0.36330735 2.08588719 0.00000000 C-CA- -0.33434665 3.29439619 -0.00119900 C-CA- -1.72917165 3.29431819 -0.00167800 C-C- -2.42677265 2.08611219 -0.00068200 H-HA- -2.27914965 -0.07418081 0.00045000 H-HA- 0.21527735 -0.07437681 0.00131500 H-HA- 0.21585335 4.24653919 -0.00125800 H-HA- -2.27929365 4.24659919 -0.00263100 C-CM- 1.90330709 2.08599922 0.00088786 H-HC- 2.43593580 2.08740001 0.92889901 C-CM- 2.58622559 2.08432974 -1.16966183 H-HC- 2.05911998 2.08292472 -2.10082116 C-C- 4.12619287 2.08445520 -1.15962425 O- 4.76677823 2.08291099 -2.24277639 S-S- 5.00306016 2.08679252 0.38940716
You can notice that actually the atom 16 which is an Oxygen had not a type assign (O- instead of O-O- or O-OH- ... it depends of your system). So you have to enter manually the type of this atom.
MM parameters missing
If you got this error message :
Generating MM parameters. Read MM parameter file: Define OH_ 1 Include all MM classes Bondstretch undefined between atoms 6 21 C-OH Angle bend undefined between atoms 1 6 21 CA-C-OH Angle bend undefined between atoms 5 6 21 CA-C-OH Angle bend undefined between atoms 23 24 25 OH-C-O2 MM function not complete Error termination via Lnk1e
That's because some of your atoms sequences are not available in the data base of AMBER, so you have to specify them by have a look at the Amber paper and take the data of some similar bonds.
You have to put the new parameters at the end of the input file and specify amber=hardfirst in the route job which correspond to "Look at the initial data base and if you do not find some data have a look at the end of the file where some are specified.
#p amber=hardfirst geom=connectivity . . . informations concerning the geometry of the system . . . HrmStr1 CM HC 367.0 1.080 HrmStr1 C S 227.0 1.810 HrmBnd1 CA CA CM 63.0 120.0 HrmBnd1 CA CM HC 35.0 123.3 HrmBnd1 CM CM HC 35.0 119.7 HrmBnd1 CM C S 50.0 120.0 HrmBnd1 HC CM C 50.0 120.0 HrmBnd1 C S CT 62.0 98.90 HrmBnd1 O C S 80.0 120.0 HrmBnd1 OH C O2 80.0 126.0
HrmStr1 enables to describe a bond length.
HrmBnd1 enables to describe an angle.
Input file problem
If you got this error message at the end of your output file:
Generating MM parameters. Read MM parameter file: End of file in RdPar. Error termination via Lnk1e
That's because you have no blank line at the end of your input file and so Gaussian is not able to read it properly. So just put some blank lines at the end of the file. Thus Gaussian is able to understand that there are no more commands at the end of the file.
If you got this one:
Read MM parameter file: Wrong or no center in RdPar. Error termination via Lnk1e
Then you may have added too few asterisks in your wildcard string e.g.: HrmBnd1 * * 0.0 0.0 rather than HrmBnd1 * * * 0.0 0.0
Conversely:
Read MM parameter file: WANTED A FLOATING POINT NUMBER AS INPUT. FOUND A STRING AS INPUT. HrmStr1 * * * 0.0 0.0
evidently means you have added too many asterisks in your wildcard string
An error message like:
Read MM parameter file: Include all MM classes Multiple matching entries: 1919 1920 1923 HrmBnd1CA C O 0.00 0.00 HrmBnd1CA C O 1.00 0.00 Inconsistent parameter file. Error termination via Lnk1e
means you've inputted 2 or more contradicting parameters for a stretch or bend. The keywords "FirstEquiv" or "LastEquiv" can be used to avoid such problems if they do occur: see the Gaussian 09 User's Reference: http://www.gaussian.com/g_tech/g_ur/k_mm.htm
Quota on your work directory : be careful !!
If your calculation stopped with no message error, but just stop during the calculation, have a look at the jobscript file corresponding to your calculation.
Concerning one of my calculation I got this "error" in the jobscript file :
Imperial College London HPC Service ----------------------------------- Job jobscript_gfp_n, jobid 2822683.cx1, username alasoro - started execution at 18:23:00 Mon 03/08/09 on system cx1-50-4-8.cx1.hpc.ic.ac.uk PGFIO/stdio: Disk quota exceeded PGFIO-F-/formatted write/unit=6/error code returned by host stdio - 122. File name = stdout formatted, sequential access record = 1 In source file getvdw.f, at line number 37 Imperial College London HPC Service ----------------------------------- Job jobscript_gfp_n, jobid 2822683.cx1, username alasoro - end of execution at 17:22:17 Tue 04/08/09 on system cx1-50-4-8.cx1.hpc.ic.ac.uk
Which correspond to : I filled in all my space available on my work directory !! yeah !!!! computational power!!!!! In my case that happen because I "accidently" generated files around 18 GB... ;-)
Some data useful concerning AMBER
How to freeze some atoms during an optimisation with ONIOM?
Use -1
When you work with a big system like a protein most of the time it is impossible to succeed an optimisation by just add Opt in the keywords, you have to run different calculations in order to optimise step by step.
Thus I had to know how to freeze some atoms during some preliminary optimisations.
When you are running an ONIOM calculation you have a column in the geometry part which is composed of 0 by default in your input file, if you change the 0 for a -1 you freeze the atom concerned.
-1 1 -1 1 -1 1 C-CA- -1 3.363213 0.007732 1.210716 H C-CA- -1 1.970023 -0.066181 1.216114 H C-CA- -1 1.273570 -0.192992 0.014410 H C-CA- -1 1.970276 -0.244749 -1.193539 H C-CA- -1 3.363106 -0.170371 -1.198894 H C-C- -1 4.059675 -0.044515 0.003339 H H-HA- -1 3.912141 0.107261 2.158300 H H-HA- -1 1.421281 -0.025737 2.168211 H H-HA- -1 1.420928 -0.344678 -2.140918 H H-HA- -1 3.912480 -0.211126 -2.150735 H C-CM- -1 -0.264208 -0.275471 0.020323 H H-HC- -1 -0.746789 -1.227926 0.089936 H C-CM- -1 -1.008342 0.854130 -0.062340 H H-HC- -1 -0.531444 1.809427 -0.132163 H C-C- -1 -2.545602 0.762540 -0.055761 H O-O- -1 -3.242820 1.807342 -0.132219 H S-S- -1 -3.338947 -0.826646 0.060392 H C-CT- -1 -5.105800 -0.611278 0.044635 H H-H1- -1 -5.582216 -1.567006 0.111806 H H-H1- -1 -5.398351 -0.130077 -0.865178 H O-OH- -1 5.487643 0.031460 -0.002449 H H-HO- 0 7.138628 -0.621023 -1.906913 L O-OH- 0 7.640691 -0.886631 -2.680855 L C-C- 0 8.416668 0.219461 -3.149170 L O-O2- 0 9.154745 0.092519 -4.160456 L C-CT- 0 8.348863 1.570623 -2.413396 L H-HC- 0 7.342784 1.934617 -2.428029 L H-HC- 0 8.666620 1.441185 -1.399899 L C-CT- 0 9.271725 2.584201 -3.115240 L H-HC- 0 8.953968 2.713639 -4.128737 L H-HC- 0 9.224614 3.522995 -2.604020 L C-CT- 0 10.719726 2.060322 -3.094181 L H-HC- 0 10.976809 1.683530 -4.062084 L
So the atoms 1 to 22 are frozen in this optimisation.
This is confirmed by the forces which are 0 for the atoms frozen.
-------------------------------------------------------------------
Center Atomic Forces (Hartrees/Bohr)
Number Number X Y Z
-------------------------------------------------------------------
1 6 0.000000000 0.000000000 0.000000000
2 6 0.000000000 0.000000000 0.000000000
3 6 0.000000000 0.000000000 0.000000000
4 6 0.000000000 0.000000000 0.000000000
5 6 0.000000000 0.000000000 0.000000000
6 6 0.000000000 0.000000000 0.000000000
7 1 0.000000000 0.000000000 0.000000000
8 1 0.000000000 0.000000000 0.000000000
9 1 0.000000000 0.000000000 0.000000000
10 1 0.000000000 0.000000000 0.000000000
11 6 0.000000000 0.000000000 0.000000000
12 1 0.000000000 0.000000000 0.000000000
13 6 0.000000000 0.000000000 0.000000000
14 1 0.000000000 0.000000000 0.000000000
15 6 0.000000000 0.000000000 0.000000000
16 8 0.000000000 0.000000000 0.000000000
17 16 0.000000000 0.000000000 0.000000000
18 6 0.000000000 0.000000000 0.000000000
19 1 0.000000000 0.000000000 0.000000000
20 1 0.000000000 0.000000000 0.000000000
21 8 0.000000000 0.000000000 0.000000000
22 1 0.000000627 0.000013720 0.000000092
23 8 -0.000007111 -0.000008031 -0.000005866
24 6 -0.000010925 -0.000000967 -0.000002673
25 8 0.000000012 0.000008672 0.000006937
26 6 -0.000010083 0.000000017 0.000003115
27 1 -0.000000709 -0.000000492 0.000000189
28 1 -0.000003917 0.000006987 0.000001626
29 6 0.000006445 -0.000003103 -0.000012735
30 1 -0.000004850 -0.000007639 -0.000006907
31 1 0.000002219 0.000011784 0.000004020
32 6 -0.000004530 -0.000000342 0.000009390
33 1 -0.000006274 -0.000005156 0.000006312
34 1 -0.000008510 -0.000003076 -0.000001552
35 1 -0.000004752 -0.000005000 -0.000000839
36 8 -0.000009012 -0.000020193 0.000017622
37 1 -0.000003546 0.000019480 -0.000016852
38 6 0.000009528 0.000011523 -0.000006439
39 6 -0.000002326 0.000007137 0.000005260
40 6 -0.000008973 0.000009163 -0.000005394
41 6 -0.000013062 -0.000018778 0.000007291
42 1 0.000012973 0.000000122 0.000018858
43 6 -0.000010444 0.000010931 -0.000008064
44 1 -0.000002307 0.000008269 -0.000004640
Use ReadFreeze
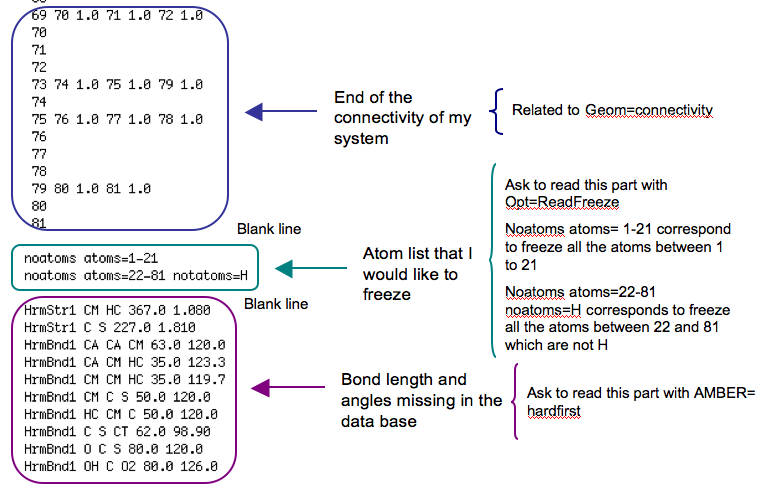
But use the command -1 could be a bit boring when you have a big system and so want to freeze some specific atoms. So if you prefer you can use an other method : ReadFreeze.
I tested Readfreeze with ONIOM. The principle is the following : the atom list, that you would like to freeze, is specified in a separate input section (terminated by a blank line) and you ask Gaussian to read this line during the optimisation with the keyword Opt=ReadFreeze in the route section.
So in the case of an optimisation with ONIOM (with QuadMacro keyword), my route section began
#p opt=(quadmacro,readfreeze) oniom(b3lyp/6-31g(d):amber=hardfirst) nosymm geom=connectivity
And I specified the atom list just after the connectivity and before the data that I specified to be able to run an AMBER calculation because my system is more an organic system than a biological system.
77 78 79 80 1.0 81 1.0 80 81 noatoms atoms=1-21 noatoms atoms=22-81 notatoms=H HrmStr1 CM HC 367.0 1.080 HrmStr1 C S 227.0 1.810 HrmBnd1 CA CA CM 63.0 120.0 HrmBnd1 CA CM HC 35.0 123.3
Nota bene
I tested Opt=ReadFreeze with many calculations : AMBER calculation, ONIOM calculation... it always works so it seems to me that it is the easier way to freeze specific atoms during an optimisation.
Back to ONIOM for biomolecules
Back to ONIOM tutorial (G03)