Zg916
NH3 molecule
Molecule name: Ammonia
N-H bond distance= 1.01798Å
H-N-H bond angle= 105.741°
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -56.55776873 au
Point group: C3v
Charge on N atom: -1.125e
Charge on H atom: 0.375e
Negative charge on N atom and positive charge on H atom are expected because N has a higher electronegativity
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
NH3 |
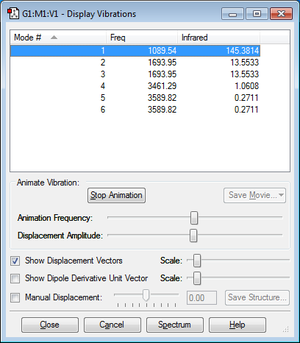
N=4,So 3N-6=6 vibrational modes are expected.
Modes 2&3 and Modes 5&6 are degenerate.
Modes 1&2&3 are bending vibrations, Modes 4&5&6 are bond stretching vibrations.
Mode 4 is highly symmetrical.
Mode 1 is known as the 'umbrella' mode.
2 bands are expected in an experimental spectrum.
H2 molecule
Molecule name: Hydrogen
H-H bond distance= 0.74279Å
H-H bond angle= 180°
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -1.17853936 au
Point group: D∞h
Vibrational frequency: 4465.68
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
N2 molecule
Molecule name: Nitrogen
N-N bond distance= 1.10550Å
N-N bond angle= 180°
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -109.52412868 au
Point group: D∞h
Vibrational frequency = 2457.33
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
E(NH3)=-56.55776873 au
2*E(NH3)=-113.1155375 au
E(N2)=-109.52412868 au
E(H2)=-1.17853936 au
3*E(H2)=-3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579074 au=-146.48 kJ/mol
Ammonia product is more stable.
Cl2 molecule
Molecule name: Chlorine
Cl-Cl bond distance= 2.04174Å1. It is quite close to the literature value which is 1.55±0.07 Å.
Cl-Cl bond angle= 180°
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -920.34987886 au
Point group: D∞h
Vibrational frequency= 520.32, it represent bond stretching
Charges on both Cl atom is 0, as they are identical
Item Value Threshold Converged? Maximum Force 0.000043 0.000450 YES RMS Force 0.000043 0.000300 YES Maximum Displacement 0.000121 0.001800 YES RMS Displacement 0.000172 0.001200 YES
Cl2 |
Some MOs
This MO is a bonding combination of the two 1s core AOs on each Cl-atom, as they are very deep in energy, which is only -101.60298 au, they do not overlap at all. As a result, this does not contribute to chemical bonding much.
This MO is a bonding combination of the two 2s AOs on each Cl-atom. Although they are much higher in energy than 1s, -9.51829 au is still very deep in energy compared to the following MOs, so they hardly overlap.
This MO is a bonding combination of the two 2px AOs on each Cl-atom which is degenerate with the antibonding combination.They overlap a little stronger than previous 2 MOs.
This MO is a bonding combination of the two 3s AOs on each Cl-atom. It is high in energy which is -0.93313 au. They overlap to form almost one surface. The energy difference between bonding and antibonding is much bigger. This is the valence orbit and contribute to the chemical bonding a lot.
This MO is a bonding combination of the two 3p AOs on each Cl-atom which is occupied. It is higher in energy than 3s combined MO, which is -0.47392 au. It contributes mainly in the chemical bonding.
Reference
1. Karan, N. K., & Arunan, E. (2004). Chlorine bond distances in ClF and Cl2 complexes. Journal of Molecular Structure, 688(1-3), 203–205. https://doi.org/10.1016/j.molstruc.2003.10.014