Yw21218
Molecule NH3
optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -56.55776873a.u.
RMS gradient: 0.00000485a.u.
Point group: C3V
Bond distance: 1.02Å
Bond angle: 106°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986263D-10
NH3 |
The optimisation file is linked to HN3 optimisation
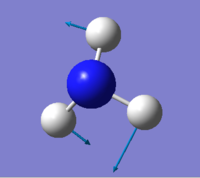
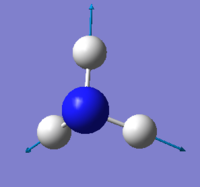
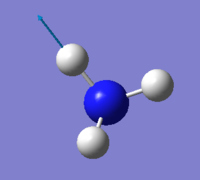
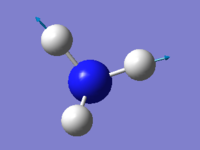
Vibration
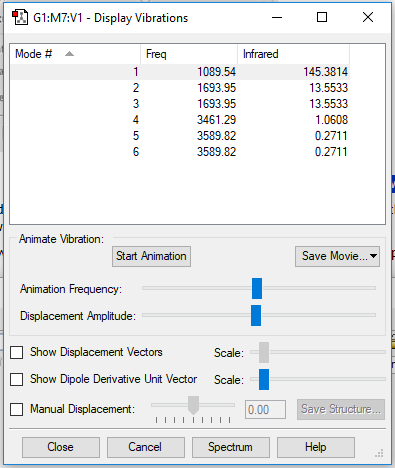
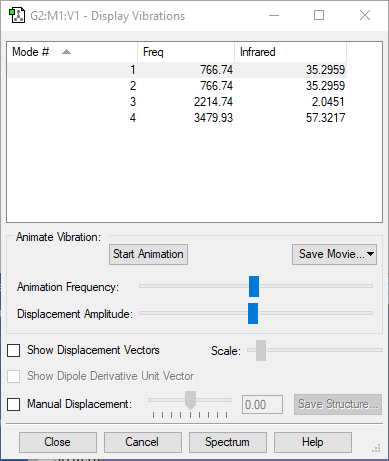
| wavenumber(cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity (arbitrary units) | 145 | 14 | 14 | 1.0 | 0.3 | 0.3 |
| image | 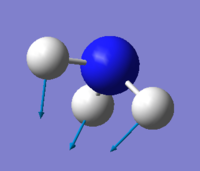 |
 |
 |
 |
 |
|
expected number of modes: 6
degenerated modes: mode 2&3, mode 5&6
bending vibration: mode 1,2,3
stretching vibration: mode 4,5,6
highly symmetric mode: mode 1&4
umbrella mode: mode 1
expected number of bands: 2
(please refer the number of mode to the screen shot attached above)
Charge
H atom: 0.472
N atom: -0.944
The results agree with the expectation, since O has higher electronegativity and pull electron density towards itself, which makes O partially negative and H partially positive. Because there are two H and one N, the amplitude of the partial negative charge on O is twicw as great as that of the positive charged H.
Molecule H2
Optimisation
Calculation method: BP3LYP
Basis set: 6-31G(d,p)
Final energy: -1.17853936 a.u.
RMS gradient: 0.00000017 a.u.
Point group: D*H
Bond distance 0.743Å
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13 Optimization completed.
H2 molecule |
The optimisation file is linked to H2 optimisation
Vibration
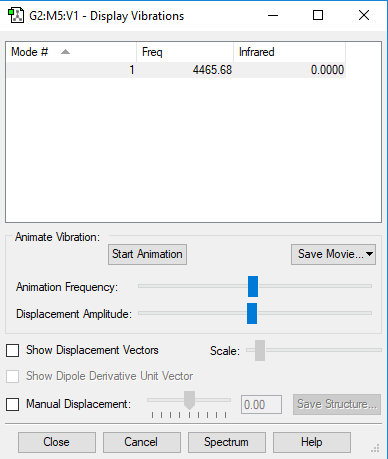
| wavenumber(cm-1) | 4466 |
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
| image | 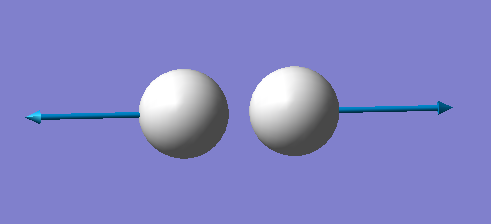
|
Charge
H atom: 0
The results agree with the expectation, it is diatomic molecule and electronegativity of both atom in the molecule is the same.
Molecule N2
Optimisation
Calculation method: BP3LYP
Basis set: 6-31G(d,p)
Final energy: -109.52412868 a.u.
RMS gradient: 0.00000060 a.u.
Point group: D*H
Bond distance 1.106 Å
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401028D-13 Optimization completed.
N2 molecule |
The optimisation file is linked to N2 optimisation
Vibration
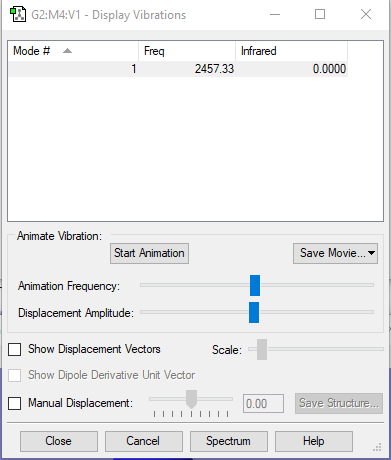
| wavenumber(cm-1) | 2457 |
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
| image | 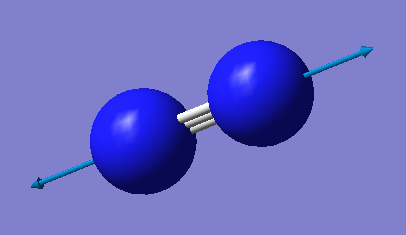
|
Charge
N atom: 0
The results agree with the expectation, it is diatomic molecule and electronegativity of both atom in the molecule is the same.
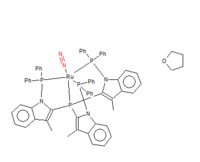
Mono-metallic transition metal complex
(dinitrogen)-{2,2',2-(phosphanetriyl)tris[1-(diphenylphosphanyl)-3-methyl-1H-indole]}-ruthenium tetrahydrofuran solvate
Unique identifier:DEKFUX
structure
(Please click the link to see more information about the molecule [[1]])
The length of nitrogen triple bond in the experimental structure is 1.086(6)Å, but the computational bond length is 1.106 Å. The experimental bond is shorter and also stronger than predicted experimental bond, because in the experimental structure N2 is a ligand that binds to a transition metal Ruthenium, which attracts the electron density of N2 and therefore shorten the bond length. This can make the ligand more tightly bind to the metal, so stabilise the structure and lower the energy of the molecule structure.
Haber-Bosch process
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.115537 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05573212 a.u. = -146.3 kJ/mol
The ΔE of the reaction is negative, which means ammonia product has lower(more negative) energy than gaseous reactants. Since the molecule with more negative energy is more stable, ammonia product is more stable in this case.
Molecule HCN
optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -93.42458132a.u.
RMS gradient: 0.00017006a.u.
Point group: C*V
CN Bond distance: 1.15702 Å
CH Bond distance: 1.06862 Å
Bond angle: 180°
Item Value Threshold Converged?
Maximum Force 0.000370 0.000450 YES
RMS Force 0.000255 0.000300 YES
Maximum Displacement 0.000676 0.001800 YES
RMS Displacement 0.000427 0.001200 YES
Predicted change in Energy=-2.062470D-07
Optimization completed.
HCN |
The optimisation file is linked to HCN optimisation
Vibration
| wavenumber(cm-1) | 767 | 767 | 2215 | 3480 |
| symmetry | PI | PI | SG | SG |
| intensity (arbitrary units) | 35 | 35 | 2.0451 | 57.3217 |
| image |  |
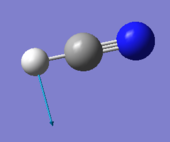 |
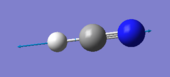 |

|
expected number of modes: 4
degenerated modes: mode 1&2
bending vibration: mode 1,2
stretching vibration: mode 3,4
expected number of bands: 2
(please refer the number of mode to the screen shot attached above)
Charge
H atom: 0.234
C atom: 0.073
N atom: 0.308
The results agree with the expectation, since N has higher electronegativity and pull electron density of C towards N, which makes N partially negative and C has smaller positive charge. As H is further away from the N, the electronegativity is not affected as much as C, so has greater positive charge on H.
Molecular Orbital

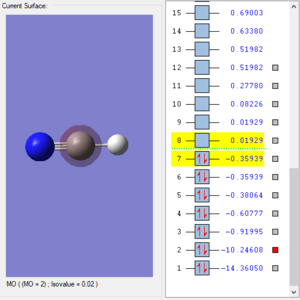
'MO 1' and 'MO 2' showed the atomic orbital of nitrogen atom 1s orbital and carbon atom 1s orbital in respectively. Since both of them are too tightly attracted by the nucleus, both of them are non-bonding orbitals and this also explains why they both have really deep energy(-14.36a.u. and -10.25 a.u. for N and C respectively). The reason why N atomic orbital has even lower energy is that N is more electronegative and bind electrons more tightly.


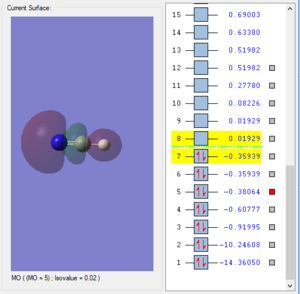

'MO 3' showed the 2s and 3s of nitrogen, 2s and 3s of carbon and 1s of hydrogen are combined together to make a bonding molecular orbital, because all of them have the same sign. Compared with the other molecular orbitals, it has the lowest energy (-0.920 a.u.). 'MO 4' showed a bonding molecular orbital between nitrogen 2s atomic orbital and mixed 2pz orbitals carbon. 'MO 5' is another bonding molecular orbital that is made up from 1s AO of H, 2p AO of C and 2p AO of N. 'MO 6' is the mixed 2p orbital contributed mainly by N and C, which forms a molecular bonding orbital. MO 6 and MO 7 have the same energy due to degenerate 2p orbitals of N and C. MO 7 is the HOMO.As you can see from the list of energy in each MO orbital, MO 3 to MO 7 have much higher energy than MO 1 and MO 2. Moreover, the energy gap becomes smaller as going up the list.

Finally, 'MO 8' shows the antibonding orbital composed of 2p orbitals of C and N. Since it is unoccupied orbital, so the energy is positive. This the LUMO. Because it is not occupied, it won't weaken the bond. Once the antibonding orbital is occupied, the bond may break.
Overall, there is no antibonding molecular orbital being occupied and each bonding orbital(MO 3 to 7) strengthen the bonds in the molecule.
Independent work:Molecule H2O
Optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Final energy: -76.41973740 a.u.
RMS gradient: 0.00006276a.u.
Point group: C2V
OH Bond distance: 0.97Å
Bond angle: 104°
Item Value Threshold Converged?
Maximum Force 0.000099 0.000450 YES
RMS Force 0.000081 0.000300 YES
Maximum Displacement 0.000128 0.001800 YES
RMS Displacement 0.000120 0.001200 YES
Predicted change in Energy=-1.939669D-08
Optimization completed.
H2O |
The optimisation file is linked to H2O optimisation
Vibration
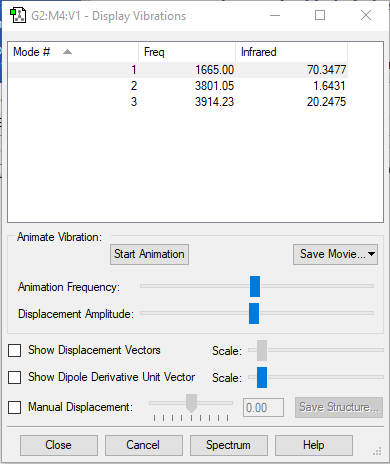
| wavenumber(cm-1) | 1665 | 3801 | 3914 |
| symmetry | A1 | A1 | B2 |
| intensity (arbitrary units) | 70 | 1.64 | 20 |
| image |  |
 |
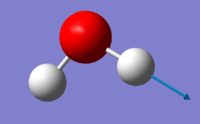
|
expected number of modes: 3
degenerated modes: N/A
bending vibration: mode 1
stretching vibration: mode 2,3
highly symmetric mode: mode 1&2
umbrella mode: mode 1
expected number of bands: 3
(please refer the number of mode to the screen shot attached above)
Charge
H atom: 0.472
O atom: -0.944
The results agree with the expectation, since O has higher electronegativity and pull electron density towards O, which makes O partially negative and H partially positive.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES - however the displayed MOs in the small molecule section are not formatted properly and how up in your independent work section.
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES - Your reported energy is wrong by 0.2 k/mol which probably is due to incorrect rounding.
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES
You analysis of the MOs is good but you missed to explicitly state the occupancy of each MO. You could have explained the relative MO energies what you have done very well for the first two ones!
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
YES - For H2O no umbrella mode exists really.
Do some deeper analysis on your results so far