Xc2017
NH3 molecules
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| Molecule name | NH3 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -56.55776873 |
| Point Group | C3V |
| N-H bond length/Å | 1.01791 |
| H-N-H bond angle | 105.741° |
NH3 |
The optimization file is linked to here
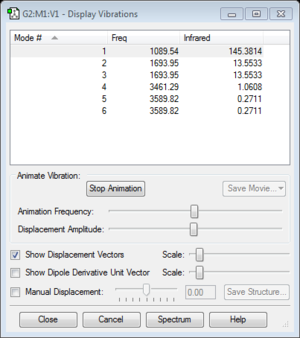
- how many modes do you expect from the 3N-6 rule? ------6
- which modes are degenerate (ie have the same energy)?------- mode 2 and 3 have same energy, mode 5 and 6 have same energy, so mode 2 and 3 ,5 and 6 are degenerate.
- which modes are "bending" vibrations and which are "bond stretch" vibrations? --------- mode 1,2 and 3 are bond bending, mode 4, 5 and 6 are bond stretch.
- which mode is highly symmetric?--------mode 4
- one mode is known as the "umbrella" mode, which one is this?-----mode 1
- how many bands would you expect to see in an experimental spectrum of gaseous ammonia?------2 bands, because mode 4, 5 and 6 are too small to be seen in the spectrum because the change in dipole moment is small, and mode 2 and 3 are degenerate, so only one band will be seen foe mode 2 and 3. Therefore, overall only 2 bands will be seen in the spectrum.
| Charge on N-atom | -1.125 |
|---|---|
| Charge on each H-atom | 0.375 |
The charge expected for N-atom is negative because N-atom has higher electronegativity than H-atom. Thus, the N-atom should be partially negative (has negative charge), and each H-atom should be partially positive. The total charge for three H-atoms is +1.125, which balanced the negative charge on N-atom and made the molecule neutral. .
N2 molecule
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------

| Molecule name | N2 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -109.52412868 |
| Point Group | D∞h |
| N-N bond length/Å | 1.10550 |
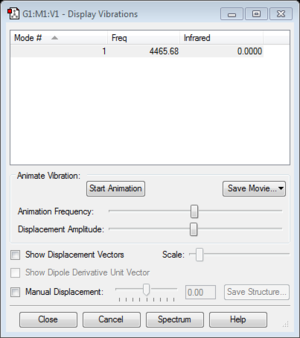
N2 |
The optimization file is lined to here
For the display vibration of N2 molecule, it can not be seen in the IR spectrum because there is no change in dipole moment when the molecule vibrate.
H2 molecules
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| Molecule name | H2 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -1.17853936 |
| Point Group | D∞h |
| H-H bond length/Å | 0.74279 |
H2 |
The optimization file is lined to here
For the display vibration of H2 molecule, it can not be seen in the IR spectrum because there is no change in dipole moment when the molecule vibrate.
Haber-Bosch reaction N2 + 3H2 -> 2NH3
- E(NH3)=-56.55776873 a.u
- 2*E(NH3)=-56.55776873*2=-113.1155375 a.u
- E(N2)=-109.52412868 a.u
- E(H2)=-1.17853936 a.u
- 3*E(H2)=-1.17853936*3=-3.53561808 a.u
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-113.1155375-(-109.52412868+(-3.53561808))=-0.05579074 a.u =-146.48 kJ/mol
The gaseous product is more stable because the value of ΔE is negative which means the product has lower energy than the reactants.
For more information see Haber process
Project molecules: CF4
Information summary
Item Value Threshold Converged?
Maximum Force 0.000078 0.000450 YES
RMS Force 0.000042 0.000300 YES
Maximum Displacement 0.000133 0.001800 YES
RMS Displacement 0.000071 0.001200 YES
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3294 -DE/DX = -0.0001 !
! R2 R(1,3) 1.3294 -DE/DX = -0.0001 !
! R3 R(1,4) 1.3294 -DE/DX = -0.0001 !
! R4 R(1,5) 1.3294 -DE/DX = -0.0001 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) -120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) 120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------

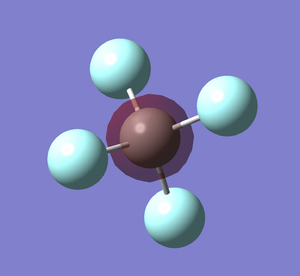
| Molecule name | CF4 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -437.47627267 |
| Point Group | Td |
| C-F bond length/Å | 1.32939 |
| F-C-F bond angle | 109.471° |
CF4 |
The optimization file is lined to here
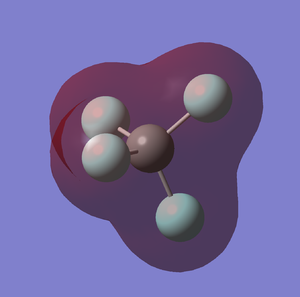
5th MO of CF4 (Figure.1)
The 5th MO (molecular orbital) is the s orbital of carbon atom, it is a non-bonding orbital which is very deep in energy (-10.52 a.u), and occupied by the 1s electrons.
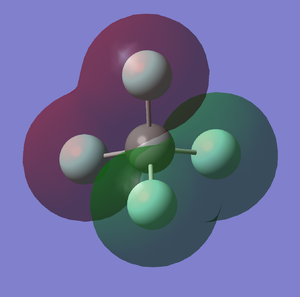
6th MO of CF4 (Figure.2)
The 6th MO (molecular orbital) is the bonding orbital consisted by the 2s orbitals of C and F atoms. It is also deep in energy (-1.37 a.u) which is much higher than the 5th MO orbital which is the non-bonding orbital. It is also an occupied orbital.
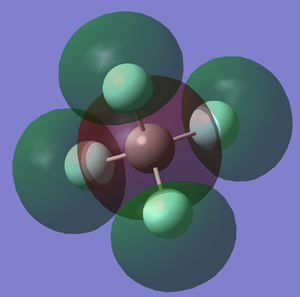
8th MO of CF4 (Figure.3)
The 8th MO (molecular orbital) is deep in energy. It is consisted by the 2p orbitals of F atom. because the C atom does not involve, this can be seen as a linkage of 2p orbital of F atoms. It has similar energy (-1.25 a.u) with the 6th MO which is the bonding orbital between s orbitals, but slightly higher in energy. It is also an occupied orbital.
10th MO of CF4 (Figure.4)
The 10th MO (molecular orbital) of CF4 atom is a bonding orbital which is also deep in energy. It is consisted by the 2p orbital of C atom and 2s orbitals of F atoms. It is also an occupied orbital.
21th MO of CF4 (Figure.5)
The 21th MO (molecular orbital) of CF4 molecule is a non-bonding orbital and it is also the HOMO (highest occupied molecular orbital) of this molecule which has energy of -0.43 a.u. It is consisted by the 2p orbitals of F atoms. It is also an occupied orbital.
Charge distribution
| charge on C atom | +1.408 |
|---|---|
| charge on each F atom | -0.352 |
The charge expected for F-atom is negative because F-atom has higher electronegativity than C-atom. Thus, each F-atom should be partially negative (has negative charge), and C-atom should be partially positive. The total charge for four F-atoms is -1.408, which balanced the positive charge on C-atom and made the molecule neutral.

Extra molecule:HCl
Information summary
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000090 0.000300 YES
Maximum Displacement 0.000139 0.001800 YES
RMS Displacement 0.000197 0.001200 YES
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.286 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------

| Molecule name | HCl |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) in a.u | -460.80077875 |
| Point Group | C∞V |
| H-Cl bond length/Å | 1.28599 |
HCl |
The optimization file is lined to here
For the display vibration of HCl molecule, it can be seen in the IR spectrum because there is a observable change in dipole moment when the molecule vibrate.
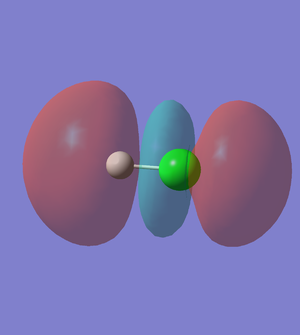
MO (molecular orbital of HCl)



5th MO of HCl (Figure.6)
The 5th MO of HCl molecule is the 2p orbital of Cl atom, and it is an occupied which is deep in energy (-7.23 a.u). The 5th MO of HCl is a non-bonding orbital.
6th MO of HCl (Figure.7)
The 6th MO of HCl is a bonding orbital consisted by the 1s orbital of H atom and the 3s orbital of Cl atom. It is also an occupied orbital which is deep in energy. However, the energy is much higher than the 5th MO which is the non-bonding orbital which is -0.85 a.u.
7th MO of HCl (Figure.8)
The 7th MO of HCl molecule is an occupied bonding orbital which is consisted by the 1s orbital of H atom and the 2p orbital of Cl atom. It is also deep in energy (-0.47 a.u).
9th MO of HCl (Figure.9)
The 9th MO of HCl molecule is the HOMO (highest occupied molecular orbital) which is a non-bonding molecular orbital. It has energy of -0.33 a.u.
10th MO of HCl (Figure.10)
It is the LUMO (lowest unoccupied molecular orbital) of the HCl molecule. It is the anti-bonding orbital which has energy of +0,.01 a.u.