XIAOJIE2012
week 1
Molecule BH3
(3-21G) Optimised BH3
test molecule |
Geometric Information
The optimised bond length(B-H)=1.19349 Å
The optimised bond angle(H-B-H)=120oc
Summary table
Real'output-Item table
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
file link
(6-31G) Optimised BH3
test molecule |
Summary table
geometric information
The optimised Bond Length B-H=1.19232 Å
The optimise H-B-H angle=120oc
Real output-Item table
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000020 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.312911D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0002 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0002 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9997 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
File link
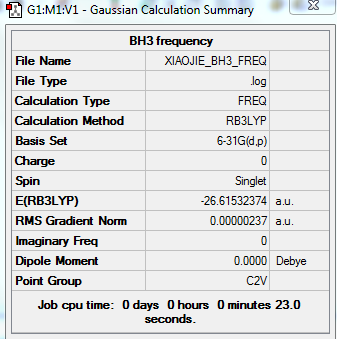
Total energies reporting
The total E=-26.46226338 a.u. for 3-21G optimised BH3
The total E=-26.61532374 a.u. for 6-31G optimised BH3
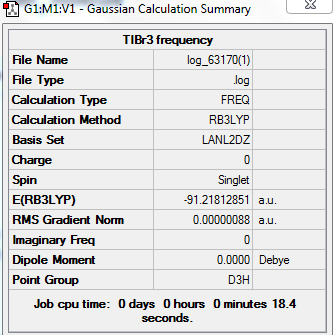
Molecule TIBr3
Optimisation TIBr3 by using LanL2DZ basis set(PPs)
test molecule |
Summary Table
'Real'output-Item table
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.083984D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Geometric Information
Bond Length TI-Br=2.65095 Å
Br-TI-Br angle=120oc
D-space link
Comparison of optimised Tl-Br bond distance and literature value
| Optimization | Literature Value | |
|---|---|---|
| Tl-Br Bond Length | 2.65095Å | 2.512Å |
The experimental value is larger than the literature value.
Molecule BBr3
Optimised BBr3 by using a mixture of basis set and PPs
test molecule |
Summary Table
Real output-Item table
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.098477D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.9339 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0022 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9956 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0022 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
File Link
Geometric Information
The optimised bond length(B-Br)=1.93394 Å
The optimised bond angle(Br-B-Br)=120.002oc
Structure Comparison
Table of the bond distances for BH3, BBr3, and TlBr3
| BH3 | BBr3 | TIBr3 |
|---|---|---|
| 1.19349 Å | 1.93394 Å | 2.65095 Å |
Bond length is the distance between centers of atoms bonded within a molecule.It depends on the size of the atoms. When we change the Ligands from H to Br, the size of the Br atom is much larger than the H atom which leads to the lengthen of the bond. Hydrogen atom Vs. Boron atom Similar: Both are small atoms Difference: B atom has five electrons while H atom only has one. In addition, the Boron atom has greater electronegativity than H. Boron atom Vs. TI atom S: They are in the same group(Periodic Table) which means they have the same number of valence electrons(3). Difference: TI atom is much bigger than the boron atom leads to the lengthen of the bond.
Question
In some structures gaussview does not draw in the bonds where we expect, does this mean there is no bond?
Ans: gaussview draws bonds based on a distance critera, so the fact that gaussview hasn't drawn bonds doesn't mean there is no bond. Just that the distance exceeds some pre-defined value. "Bonds" in gausview are a structural convenience.
What is a bond
Ans: A bond is an attraction between atoms. Atoms share their electrons to form the bond.(achieve 8 e- =octet rule)
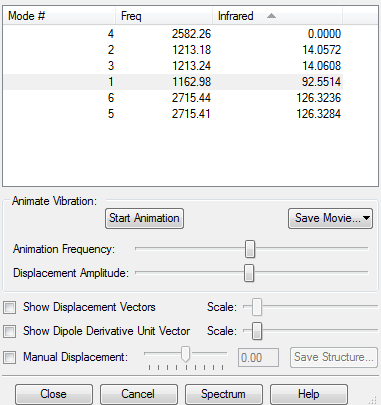
Frequency Analysis for BH3
Summary Table
low frequencies for the BH3 molecule
Low frequencies --- -18.6669 -0.0009 -0.0003 0.0006 12.5167 12.5631 Low frequencies --- 1162.9785 1213.1756 1213.2363
Frequency file link
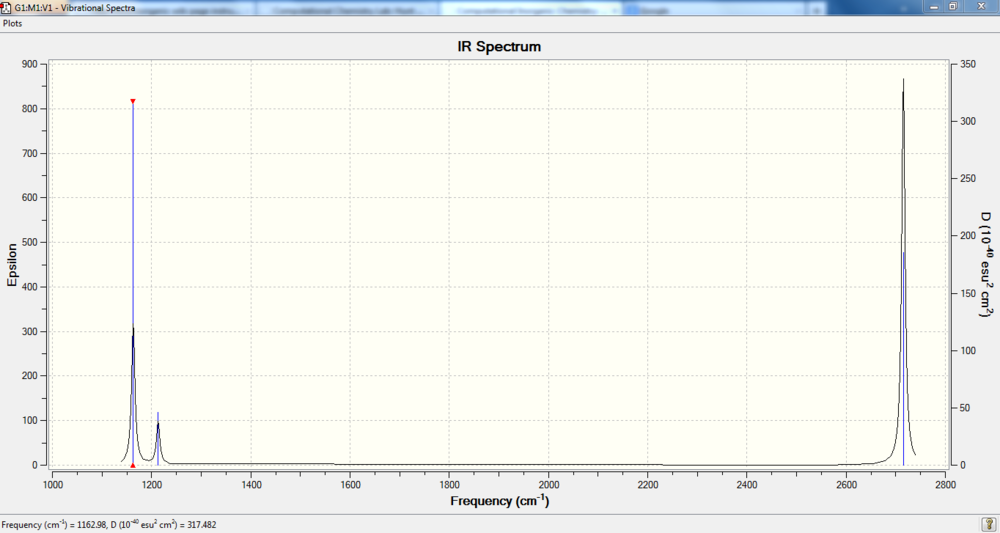
Vibrational Analysis For BH3
IR Spectrum
IR Spectrum Analysis:
The value under the IR indicates the intensity of the peak. It is clear that Mode 4 has zero intensity which leads to a missing peak in the spectrum. In addition ,there are two pair vibrations with similar intensity. So these 2 pair vibration modes combine to give 2 peaks. The last one goes to mode .
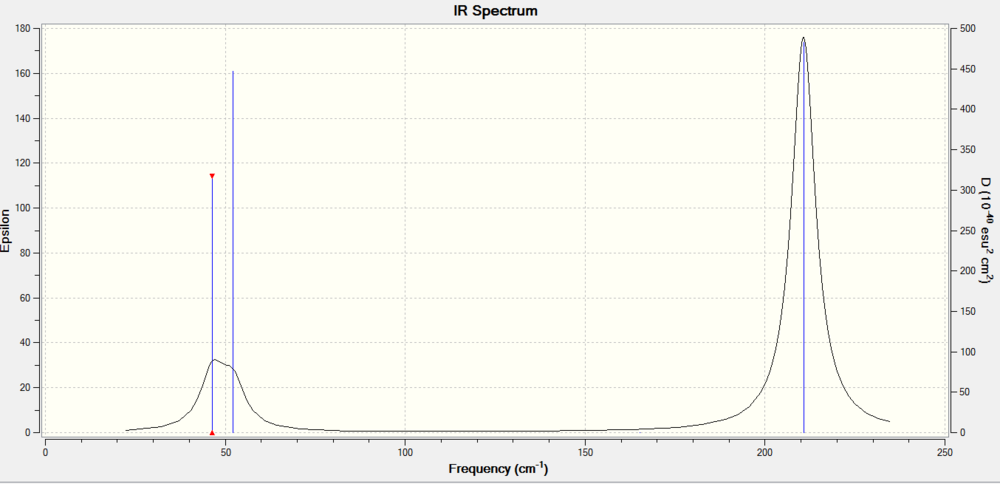
Frequency Analysis for TlBr3
Summary table
Low frequency
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
D-space link
IR Spectrum
Comparison of the Vibration frequency of BH3 AND TIBr3
| BH3 | TIBr3 |
|---|---|
| 1162.98 | 46.43 |
| 1213.18 | 46.43 |
| 1213.24 | 52.14 |
| 2582.26 | 165.27 |
| 2715.41 | 210.69 |
| 2715.44 | 210.69 |
The difference between frequencies of BH3 and TIBr3 is large. TI-Br bond is much weaker than the B-H bond(long bond distance,weaker bonds) which leads to the low frequencies. By animating the vibrations , we can find that both two molecules have 6 mode vibrations and the number of the peaks are the same(3). In additon,there has been a reordering of modes. For BH3 molecule, mode1( A2' ) has the lowest frequency. while in TIBr3 molecule, the two degenerate modes (1 and 2) have the lowest frequency.
Question
Why must you use the same method and basis set for both the optimisation and frequency analysis calculations?
The basis set determines the accuracy, and the total energy for any calculation is highly dependent on the quality of the basis set. So we can never use different basis set for analysis.
What is the purpose of carrying out a frequency analysis?
The frequency analysis is the second derivative of the potential energy surface, if the frequencies are all positive then we have a minimum, if one of them is negative we have a transition state. The frequency analysis is important because it provides the IR and Raman modes to compare with experiment.
What do the "Low frequencies" represent?
We need to make sure there is no negative value which means that we have failed to find a critical point and the optimisation has not completed or has failed.
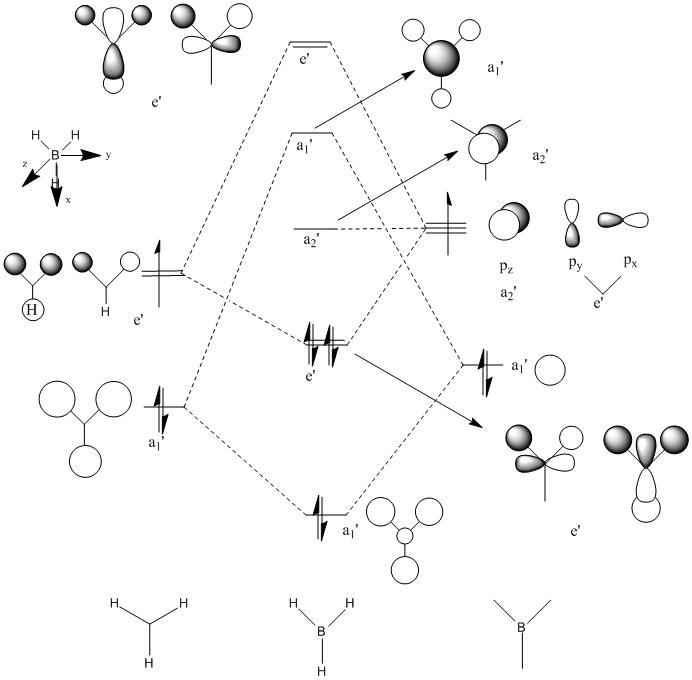
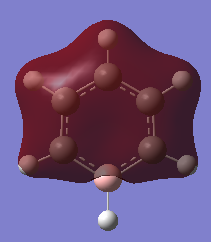
Molecule Orbitals of BH3
D-space link
MO Diagram Chemdraw
Real MOs
 |
 |
 |

|
| a1' | e' | e' | a2' |
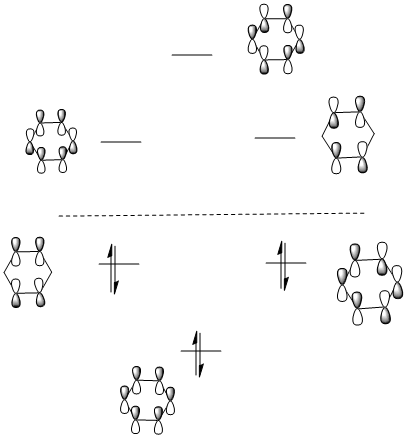
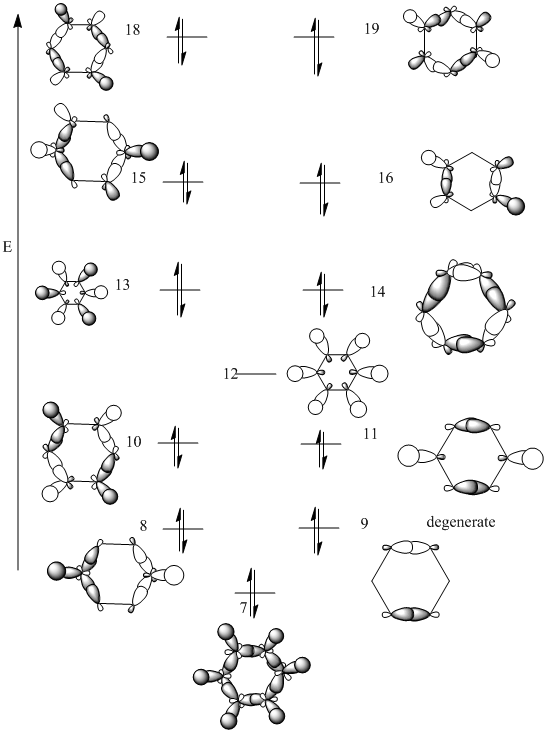
The calculated molecular orbitals match up with the LCAO MOs. And the computed MOs is much easier to find the key information,such as atom contribution to the MO, presence of the nodes.
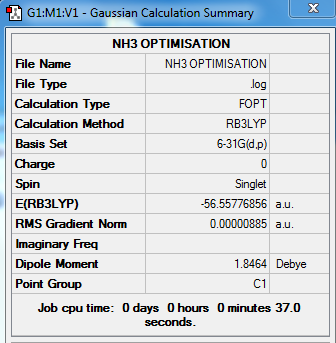
Molecule NH3
6-31G Optimised NH3
test molecule |
Summary Table
Real output--Item table
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629731D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Geometric Information
Bond Length N-H=1.01797 Å
H-N-H angle=105.741oc
Frequency Analysis for NH3
summary table
low frequency
Low frequencies --- -30.6965 -0.0012 0.0012 0.0015 20.2203 28.2940 Low frequencies --- 1089.5540 1694.1243 1694.1861
frequency file link
MO analysis For NH3
File Link
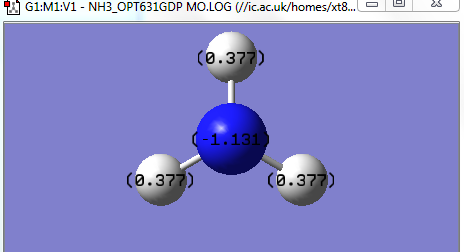
NBO Analysis for NH3
Charge Distribution and charge number
Real output-file
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.13121 1.99981 6.11785 0.01356 8.13121
H 2 0.37707 0.00000 0.62078 0.00214 0.62293
H 3 0.37707 0.00000 0.62078 0.00214 0.62293
H 4 0.37707 0.00000 0.62078 0.00214 0.62293
=======================================================================
* Total * 0.00000 1.99981 7.98020 0.01999 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99945) BD ( 1) N 1 - H 2
( 68.90%) 0.8301* N 1 s( 26.41%)p 2.78( 73.48%)d 0.00( 0.11%)
0.0001 0.5139 0.0035 0.0000 0.0000
0.0000 0.8154 0.0267 -0.2630 0.0052
0.0000 0.0000 -0.0295 -0.0146 -0.0008
( 31.10%) 0.5576* H 2 s( 99.85%)p 0.00( 0.15%)
0.9992 0.0001 0.0000 -0.0379 0.0101
2. (1.99945) BD ( 1) N 1 - H 3
( 68.90%) 0.8301* N 1 s( 26.41%)p 2.78( 73.48%)d 0.00( 0.11%)
0.0001 0.5139 0.0035 0.0000 -0.7062
-0.0231 -0.4077 -0.0134 -0.2630 0.0052
0.0126 0.0255 0.0147 0.0073 -0.0008
( 31.10%) 0.5576* H 3 s( 99.85%)p 0.00( 0.15%)
0.9992 0.0001 0.0328 0.0190 0.0101
3. (1.99945) BD ( 1) N 1 - H 4
( 68.90%) 0.8301* N 1 s( 26.41%)p 2.78( 73.48%)d 0.00( 0.11%)
0.0001 0.5139 0.0035 0.0000 0.7062
0.0231 -0.4077 -0.0134 -0.2630 0.0052
-0.0126 -0.0255 0.0147 0.0073 -0.0008
( 31.10%) 0.5576* H 4 s( 99.85%)p 0.00( 0.15%)
0.9992 0.0001 -0.0328 0.0190 0.0101
4. (1.99981) CR ( 1) N 1 s(100.00%)
1.0000 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99826) LP ( 1) N 1 s( 20.74%)p 3.82( 79.19%)d 0.00( 0.07%)
0.0001 0.4553 -0.0082 0.0000 0.0000
0.0000 0.0000 0.0000 0.8886 -0.0476
0.0000 0.0000 0.0000 0.0000 -0.0256
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99945 -0.82283
2. BD ( 1) N 1 - H 3 1.99945 -0.82283
3. BD ( 1) N 1 - H 4 1.99945 -0.82283
4. CR ( 1) N 1 1.99981 -15.38421
5. LP ( 1) N 1 1.99826 -0.48521 16(v),20(v),24(v)
Association energies: Ammonia-Borane
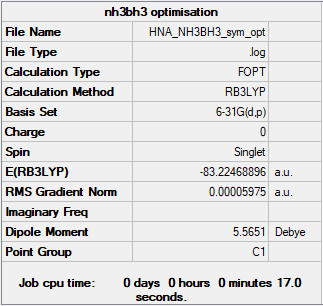
Optimisation of NH3BH3 by using b3lyp/6-31G(d,p) level
test molecule |
Summary Table
Real output-Item table
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000606 0.001800 YES
RMS Displacement 0.000336 0.001200 YES
Predicted change in Energy=-1.993999D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,8) 1.2101 -DE/DX = -0.0001 !
! R5 R(5,8) 1.2101 -DE/DX = -0.0001 !
! R6 R(6,8) 1.2101 -DE/DX = -0.0001 !
! R7 R(7,8) 1.668 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.87 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.8652 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0329 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.8697 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0286 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0291 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.8693 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8721 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.6003 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.8747 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.6003 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.5984 -DE/DX = 0.0 !
! D1 D(1,7,8,4) -179.9867 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9892 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0135 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9839 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0136 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -179.9837 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0161 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9864 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.9837 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
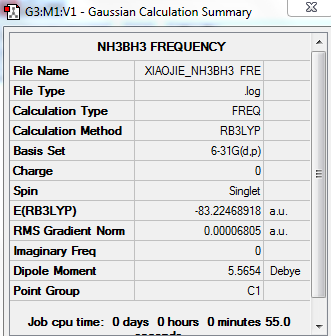
Frequency analysis of NH3BH3
Summary table
Low frequency
Low frequencies --- -0.0010 0.0002 0.0012 17.1579 22.5687 38.9093 Low frequencies --- 265.8799 632.3786 639.0712
Frequency file link
ENERGY DIFFERENCE
E(NH3)=-56.55776856 a.u.
E(BH3)=-26.61532374 a.u.
E(NH3BH3)=-83.22468918 a.u.
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
=-83.22468918-[(-56.55776856)+(-26.61532374)] =-0.0516 a.u. =-135.4758KJ/mol
Mini Project--Aromaticity
Benzene
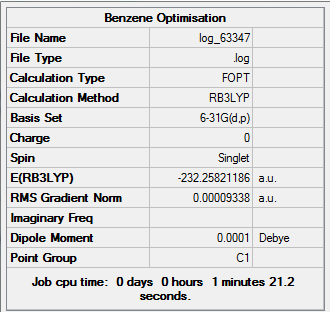
(6-31G)optimised Benzene
test molecule |
Summary Table
Real Output-Item table
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000084 0.000300 YES
Maximum Displacement 0.000870 0.001800 YES
RMS Displacement 0.000313 0.001200 YES
Predicted change in Energy=-4.983462D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.0861 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.0861 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9996 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9968 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0036 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0036 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9916 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0048 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9967 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0101 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9932 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9996 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9891 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0113 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.004 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0048 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9912 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9965 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0072 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9963 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0021 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.0099 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0016 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.0068 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0119 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0087 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0039 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0007 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0064 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0058 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9968 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0026 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.005 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0061 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0057 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0055 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0085 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradG
D-space link [11]
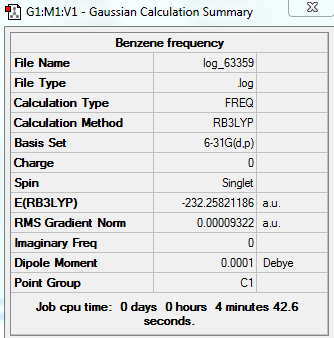
Frequency Analysis for Benzene
Summary Table
Low Frequency
Low frequencies --- -14.2145 -2.6928 -0.0007 -0.0005 0.0009 10.0061 Low frequencies --- 413.7279 414.5534 621.0446
D space Link
MO Analysis for Benzene
D space Link
MO diagram
NBO Analysis for Benzene
Charge Distribution
Charge Number
Real Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.23856 1.99910 4.22615 0.01331 6.23856
C 2 -0.23855 1.99910 4.22613 0.01331 6.23855
C 3 -0.23853 1.99910 4.22612 0.01331 6.23853
C 4 -0.23856 1.99910 4.22615 0.01331 6.23856
C 5 -0.23854 1.99910 4.22613 0.01331 6.23854
C 6 -0.23853 1.99910 4.22612 0.01331 6.23853
H 7 0.23855 0.00000 0.76001 0.00144 0.76145
H 8 0.23854 0.00000 0.76002 0.00144 0.76146
H 9 0.23855 0.00000 0.76002 0.00144 0.76145
H 10 0.23855 0.00000 0.76001 0.00144 0.76145
H 11 0.23854 0.00000 0.76002 0.00144 0.76146
H 12 0.23855 0.00000 0.76002 0.00144 0.76145
=======================================================================
* Total * 0.00000 11.99462 29.91692 0.08846 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98096) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.7709
-0.0075 -0.2281 -0.0349 -0.0003 0.0000
0.0069 0.0000 0.0000 0.0151 -0.0109
( 50.00%) 0.7071* C 2 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.7816
0.0240 0.1884 -0.0264 0.0002 0.0000
0.0096 0.0000 0.0000 0.0135 -0.0109
2. (1.98097) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.2276
0.0349 0.7710 0.0076 0.0006 0.0000
0.0069 0.0000 0.0000 -0.0151 -0.0109
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.1878
0.0264 -0.7817 -0.0240 -0.0006 0.0000
0.0096 0.0000 0.0000 -0.0135 -0.0109
3. (1.66532) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0052 0.0188 0.0000 0.0000
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0139 -0.0137 0.0000 0.0000
4. (1.98305) BD ( 1) C 1 - H 7
( 62.04%) 0.7876* C 1 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.5933
-0.0103 -0.5930 0.0103 -0.0003 0.0000
-0.0166 0.0000 0.0000 0.0000 -0.0105
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0161 0.0161 0.0000
5. (1.98097) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.5830
-0.0340 0.5535 -0.0109 0.0004 0.0000
-0.0165 0.0000 0.0000 -0.0015 -0.0109
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.5539
-0.0109 -0.5826 -0.0340 -0.0004 0.0000
-0.0165 0.0000 0.0000 0.0015 -0.0109
6. (1.66532) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0049 0.0189 0.0000 0.0000
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 0.0189 -0.0049 0.0000 0.0000
7. (1.98305) BD ( 1) C 2 - H 8
( 62.04%) 0.7876* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.2167
-0.0038 0.8104 -0.0141 0.0006 0.0000
-0.0083 0.0000 0.0000 0.0144 0.0105
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0059 -0.0220 0.0000
8. (1.98096) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.1879
-0.0264 0.7817 0.0240 0.0006 0.0000
0.0096 0.0000 0.0000 -0.0135 -0.0109
( 50.00%) 0.7071* C 4 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.2276
-0.0349 -0.7711 -0.0076 -0.0005 0.0000
0.0069 0.0000 0.0000 -0.0151 -0.0109
9. (1.98305) BD ( 1) C 3 - H 9
( 62.04%) 0.7876* C 3 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.8102
-0.0141 0.2173 -0.0038 0.0002 0.0000
-0.0083 0.0000 0.0000 -0.0144 0.0105
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0220 -0.0059 0.0000
10. (1.98097) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.7709
0.0076 0.2282 0.0349 0.0002 0.0000
0.0069 0.0000 0.0000 0.0151 -0.0109
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.7815
-0.0240 -0.1884 0.0264 -0.0003 0.0000
0.0096 0.0000 0.0000 0.0135 -0.0109
11. (1.66534) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 0.0188 -0.0052 0.0000 0.0000
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0137 -0.0139 0.0000 0.0000
12. (1.98305) BD ( 1) C 4 - H 10
( 62.04%) 0.7876* C 4 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.5934
-0.0103 -0.5929 0.0103 -0.0003 0.0000
0.0166 0.0000 0.0000 0.0000 0.0105
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0161 0.0161 0.0000
13. (1.98096) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.5831
0.0340 -0.5535 0.0109 -0.0003 0.0000
-0.0165 0.0000 0.0000 -0.0015 -0.0109
( 50.00%) 0.7071* C 6 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.5540
0.0109 0.5826 0.0340 0.0003 0.0000
-0.0165 0.0000 0.0000 0.0015 -0.0109
14. (1.98305) BD ( 1) C 5 - H 11
( 62.04%) 0.7876* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.2169
-0.0038 0.8103 -0.0141 0.0006 0.0000
0.0083 0.0000 0.0000 -0.0144 -0.0105
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0059 -0.0220 0.0000
15. (1.98305) BD ( 1) C 6 - H 12
( 62.04%) 0.7876* C 6 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.8102
-0.0141 0.2175 -0.0038 0.0003 0.0000
0.0083 0.0000 0.0000 0.0144 -0.0105
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0220 -0.0059 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98096 -0.68186 110(g),107(g),114(v),120(v)
73(v),43(v),109(g),112(g)
42(v),72(v)
2. BD ( 1) C 1 - C 6 1.98097 -0.68200 118(g),106(g),119(v),112(v)
33(v),63(v),109(g),120(g)
62(v),32(v)
3. BD ( 2) C 1 - C 6 1.66532 -0.23794 111(v),116(v),35(v),65(v)
4. BD ( 1) C 1 - H 7 1.98305 -0.51236 118(v),110(v),72(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98097 -0.68202 106(g),113(g),109(v),117(v)
23(v),53(v),114(g),112(g)
52(v),22(v)
6. BD ( 2) C 2 - C 3 1.66532 -0.23794 108(v),116(v),55(v),25(v)
7. BD ( 1) C 2 - H 8 1.98305 -0.51234 113(v),107(v),42(v),22(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98096 -0.68184 110(g),115(g),112(v),119(v)
63(v),33(v),114(g),117(g)
32(v),62(v)
9. BD ( 1) C 3 - H 9 1.98305 -0.51236 106(v),115(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - C 5 1.98097 -0.68202 118(g),113(g),114(v),120(v)
43(v),73(v),117(g),119(g)
72(v),42(v)
11. BD ( 2) C 4 - C 5 1.66534 -0.23795 108(v),111(v),45(v),75(v)
12. BD ( 1) C 4 - H 10 1.98305 -0.51236 118(v),110(v),62(v),42(v)
115(g),113(g)
13. BD ( 1) C 5 - C 6 1.98096 -0.68186 115(g),107(g),109(v),117(v)
53(v),23(v),119(g),120(g)
22(v),52(v)
14. BD ( 1) C 5 - H 11 1.98305 -0.51234 113(v),107(v),52(v),72(v)
115(g),118(g)
15. BD ( 1) C 6 - H 12 1.98305 -0.51236 106(v),115(v),22(v),62(v)
107(g),118(g)
16. CR ( 1) C 1 1.99911 -10.04057 73(v),33(v),110(v),118(v)
120(v),112(v)
17. CR ( 1) C 2 1.99911 -10.04056 43(v),23(v),113(v),107(v)
114(v),109(v)
18. CR ( 1) C 3 1.99911 -10.04057 33(v),53(v),106(v),115(v)
112(v),117(v)
19. CR ( 1) C 4 1.99911 -10.04057 63(v),43(v),118(v),110(v)
119(v),114(v)
20. CR ( 1) C 5 1.99911 -10.04056 53(v),73(v),113(v),107(v)
117(v),120(v)
21. CR ( 1) C 6 1.99911 -10.04057 23(v),63(v),106(v),115(v)
109(v),119(v)
discussion
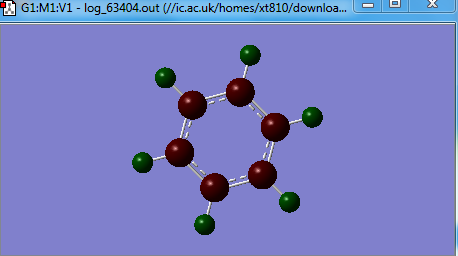
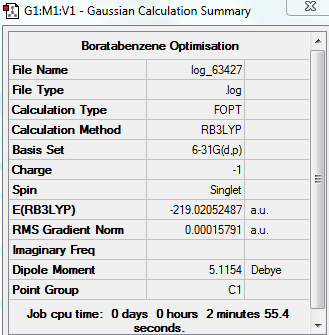
Boratabenzne
OPtimised Boratabenzne
test molecule |
Summary Table
Real Output-Item Table
Item Value Threshold Converged?
Maximum Force 0.000160 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000938 0.001800 YES
RMS Displacement 0.000327 0.001200 YES
Predicted change in Energy=-6.588035D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3989 -DE/DX = 0.0 !
! R2 R(1,7) 1.097 -DE/DX = -0.0001 !
! R3 R(1,12) 1.5138 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,8) 1.0968 -DE/DX = 0.0001 !
! R6 R(3,4) 1.4053 -DE/DX = -0.0001 !
! R7 R(3,9) 1.0917 -DE/DX = -0.0001 !
! R8 R(4,5) 1.3989 -DE/DX = 0.0 !
! R9 R(4,10) 1.0968 -DE/DX = 0.0001 !
! R10 R(5,11) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5137 -DE/DX = 0.0001 !
! R12 R(6,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,7) 115.9492 -DE/DX = 0.0001 !
! A2 A(2,1,12) 120.0804 -DE/DX = -0.0001 !
! A3 A(7,1,12) 123.9704 -DE/DX = -0.0001 !
! A4 A(1,2,3) 122.1345 -DE/DX = 0.0001 !
! A5 A(1,2,8) 120.4329 -DE/DX = -0.0002 !
! A6 A(3,2,8) 117.4326 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.4538 -DE/DX = -0.0001 !
! A8 A(2,3,9) 119.7747 -DE/DX = 0.0001 !
! A9 A(4,3,9) 119.7715 -DE/DX = 0.0001 !
! A10 A(3,4,5) 122.138 -DE/DX = 0.0001 !
! A11 A(3,4,10) 117.4424 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.4196 -DE/DX = -0.0002 !
! A13 A(4,5,11) 115.9544 -DE/DX = 0.0001 !
! A14 A(4,5,12) 120.0799 -DE/DX = -0.0001 !
! A15 A(11,5,12) 123.9657 -DE/DX = -0.0001 !
! A16 A(1,12,5) 115.1135 -DE/DX = 0.0 !
! A17 A(1,12,6) 122.4375 -DE/DX = 0.0 !
! A18 A(5,12,6) 122.449 -DE/DX = 0.0 !
! D1 D(7,1,2,3) -180.0017 -DE/DX = 0.0 !
! D2 D(7,1,2,8) 0.0007 -DE/DX = 0.0 !
! D3 D(12,1,2,3) -0.0023 -DE/DX = 0.0 !
! D4 D(12,1,2,8) 180.0001 -DE/DX = 0.0 !
! D5 D(2,1,12,5) 0.0013 -DE/DX = 0.0 !
! D6 D(2,1,12,6) 180.001 -DE/DX = 0.0 !
! D7 D(7,1,12,5) 180.0006 -DE/DX = 0.0 !
! D8 D(7,1,12,6) 0.0002 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.9992 -DE/DX = 0.0 !
! D11 D(8,2,3,4) -180.0023 -DE/DX = 0.0 !
! D12 D(8,2,3,9) -0.0015 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0035 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0018 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 180.0027 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0026 -DE/DX = 0.0 !
! D17 D(3,4,5,11) -180.0044 -DE/DX = 0.0 !
! D18 D(3,4,5,12) -0.0044 -DE/DX = 0.0 !
! D19 D(10,4,5,11) 0.001 -DE/DX = 0.0 !
! D20 D(10,4,5,12) 180.0011 -DE/DX = 0.0 !
! D21 D(4,5,12,1) 0.002 -DE/DX = 0.0 !
! D22 D(4,5,12,6) 180.0023 -DE/DX = 0.0 !
! D23 D(11,5,12,1) 180.002 -DE/DX = 0.0 !
! D24 D(11,5,12,6) 0.0023 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-space Link
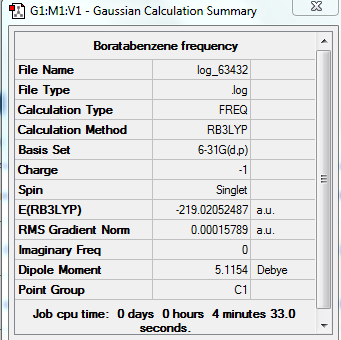
Frequency Analysis For Boratabenzne
Summary Table
Low Frequency
Low frequencies --- -13.9534 -0.0007 -0.0006 -0.0005 9.6265 14.6141 Low frequencies --- 371.0129 404.6537 565.1741
D-space Link
MO Analysis for Boratabenzene
D-space Link
NBO Anylysis for Boratabenzene
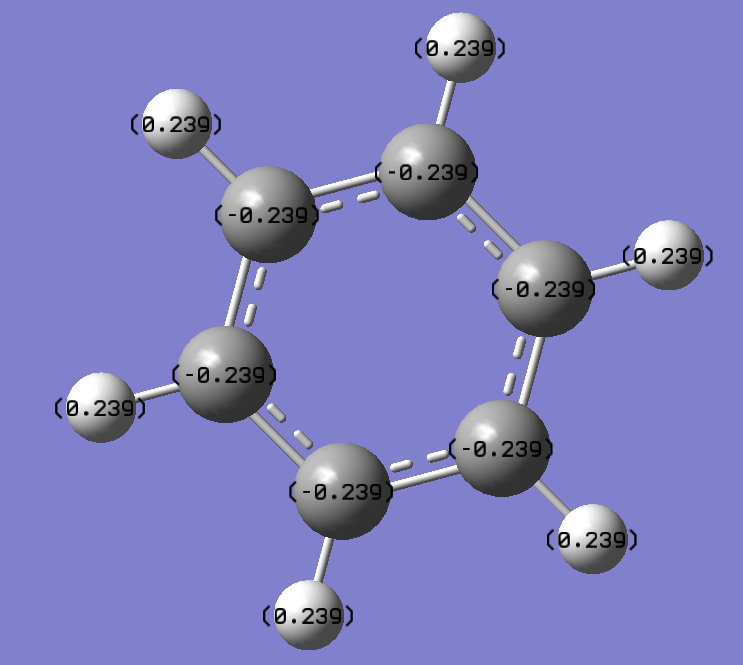
Charge Distribution
Charge Number
Real Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.58790 1.99901 4.57711 0.01178 6.58790
C 2 -0.25043 1.99910 4.23720 0.01412 6.25043
C 3 -0.33983 1.99907 4.32693 0.01384 6.33983
C 4 -0.25042 1.99910 4.23719 0.01412 6.25042
C 5 -0.58793 1.99901 4.57714 0.01178 6.58793
H 6 -0.09642 0.00000 1.09588 0.00054 1.09642
H 7 0.18380 0.00000 0.81402 0.00218 0.81620
H 8 0.17906 0.00000 0.81831 0.00262 0.82094
H 9 0.18563 0.00000 0.81237 0.00200 0.81437
H 10 0.17906 0.00000 0.81832 0.00262 0.82094
H 11 0.18380 0.00000 0.81402 0.00218 0.81620
B 12 0.20157 1.99906 2.78777 0.01160 4.79843
=======================================================================
* Total * -1.00000 11.99436 29.91625 0.08939 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98270) BD ( 1) C 1 - C 2
( 49.23%) 0.7017* C 1 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5696 -0.0200 0.0010 0.0024
-0.0269 -0.8202 -0.0353 -0.0003 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
( 50.77%) 0.7125* C 2 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0573
-0.0311 0.7869 0.0164 0.0003 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
2. (1.76864) BD ( 2) C 1 - C 2
( 51.87%) 0.7202* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0006
0.0000 -0.0004 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
( 48.13%) 0.6938* C 2 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0006
0.0000 -0.0004 0.0000 0.9996 -0.0214
0.0000 0.0031 0.0171 0.0000 0.0000
3. (1.98420) BD ( 1) C 1 - H 7
( 59.41%) 0.7708* C 1 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7906
0.0003 0.3472 0.0088 0.0006 0.0000
-0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 7 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
4. (1.96996) BD ( 1) C 1 - B 12
( 66.70%) 0.8167* C 1 s( 42.04%)p 1.38( 57.95%)d 0.00( 0.01%)
0.0000 0.6482 0.0158 0.0012 0.6117
-0.0293 0.4521 0.0090 -0.0002 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7057
-0.0393 -0.4069 0.0096 0.0002 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
5. (1.97970) BD ( 1) C 2 - C 3
( 49.96%) 0.7068* C 2 s( 35.50%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6874
0.0034 -0.4135 -0.0325 -0.0006 0.0000
-0.0146 0.0000 0.0000 0.0080 -0.0107
( 50.04%) 0.7074* C 3 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7061
-0.0327 0.3754 -0.0141 0.0006 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
6. (1.98570) BD ( 1) C 2 - H 8
( 59.32%) 0.7702* C 2 s( 26.88%)p 2.72( 73.07%)d 0.00( 0.05%)
0.0003 -0.5183 -0.0133 0.0012 0.7229
-0.0089 0.4561 -0.0100 -0.0002 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
7. (1.97971) BD ( 1) C 3 - C 4
( 50.04%) 0.7074* C 3 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7063
0.0327 0.3752 -0.0141 -0.0003 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 4 s( 35.51%)p 1.82( 64.45%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6875
-0.0034 -0.4132 -0.0325 0.0003 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
8. (1.98507) BD ( 1) C 3 - H 9
( 59.44%) 0.7710* C 3 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 0.0002
0.0000 0.8469 -0.0076 0.0003 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 9 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
9. (1.98270) BD ( 1) C 4 - C 5
( 50.77%) 0.7125* C 4 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0576
0.0311 0.7869 0.0164 0.0003 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 5 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0028
0.0269 -0.8202 -0.0353 -0.0003 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
10. (1.76861) BD ( 2) C 4 - C 5
( 48.13%) 0.6937* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0006
0.0000 -0.0004 0.0000 0.9996 -0.0214
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0006
0.0000 -0.0004 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
11. (1.98570) BD ( 1) C 4 - H 10
( 59.32%) 0.7702* C 4 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7227
-0.0089 -0.4564 0.0100 -0.0006 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 10 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
12. (1.98420) BD ( 1) C 5 - H 11
( 59.41%) 0.7708* C 5 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7907
-0.0003 0.3469 0.0088 -0.0003 0.0000
0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
13. (1.96996) BD ( 1) C 5 - B 12
( 66.70%) 0.8167* C 5 s( 42.04%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6482 -0.0158 -0.0012 0.6115
-0.0293 -0.4524 -0.0090 -0.0005 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7055
-0.0393 0.4072 -0.0096 0.0006 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
14. (1.98604) BD ( 1) H 6 - B 12
( 55.09%) 0.7422* H 6 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 0.0002
0.0000 0.8172 -0.0016 0.0003 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.98270 -0.46493 110(g),111(g),114(v),112(g)
44(v),120(v),109(g),98(v)
43(v)
2. BD ( 2) C 1 - C 2 1.76864 -0.02906 21(v),22(v),45(v),100(v)
108(g)
3. BD ( 1) C 1 - H 7 1.98420 -0.28848 111(v),110(g),33(v),119(v)
97(v),107(g)
4. BD ( 1) C 1 - B 12 1.96996 -0.31778 107(g),112(v),118(v),109(g)
34(v),33(v),77(v),64(v)
119(g)
5. BD ( 1) C 2 - C 3 1.97970 -0.46973 107(g),113(g),117(v),109(v)
54(v),23(v),114(g),112(g)
6. BD ( 1) C 2 - H 8 1.98570 -0.31414 113(v),110(v),43(v),23(v)
107(g)
7. BD ( 1) C 3 - C 4 1.97971 -0.46977 115(g),111(g),112(v),118(v)
34(v),63(v),114(g),117(g)
8. BD ( 1) C 3 - H 9 1.98507 -0.31743 115(v),107(v),53(v),33(v)
113(g),111(g)
9. BD ( 1) C 4 - C 5 1.98270 -0.46494 119(g),113(g),114(v),117(g)
44(v),120(v),118(g),98(v)
43(v)
10. BD ( 2) C 4 - C 5 1.76861 -0.02906 21(v),22(v),45(v),100(v)
116(g)
11. BD ( 1) C 4 - H 10 1.98570 -0.31413 111(v),119(v),43(v),63(v)
115(g)
12. BD ( 1) C 5 - H 11 1.98420 -0.28848 113(v),119(g),53(v),110(v)
97(v),115(g)
13. BD ( 1) C 5 - B 12 1.96996 -0.31781 115(g),117(v),109(v),118(g)
54(v),53(v),93(v),24(v)
110(g)
14. BD ( 1) H 6 - B 12 1.98604 -0.17251 107(v),115(v),23(v),63(v)
15. CR ( 1) C 1 1.99902 -9.79409 34(v),98(v),110(g),111(v)
112(v),97(v)
16. CR ( 1) C 2 1.99910 -9.83480 24(v),44(v),110(v),114(v)
113(v),109(v)
17. CR ( 1) C 3 1.99907 -9.82827 34(v),54(v),115(v),107(v)
117(v),112(v),37(v),57(v)
53(v),33(v)
18. CR ( 1) C 4 1.99910 -9.83478 64(v),44(v),119(v),114(v)
111(v),118(v)
19. CR ( 1) C 5 1.99902 -9.79407 54(v),98(v),119(g),113(v)
117(v),97(v)
20. CR ( 1) B 12 1.99907 -6.36939 109(v),118(v),115(v),107(v)
63(v),23(v)
21. LP ( 1) C 3 1.14687 0.09689 116(v),108(v),45(g),56(v)
36(v),35(v),55(v)
22. LP*( 1) B 12 0.57264 0.22268 108(v),116(v),100(g),67(v)
27(v)
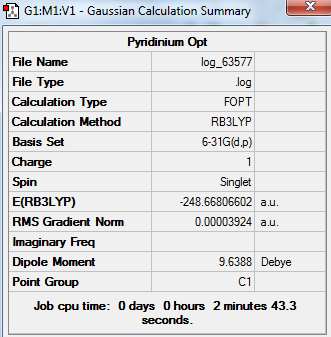
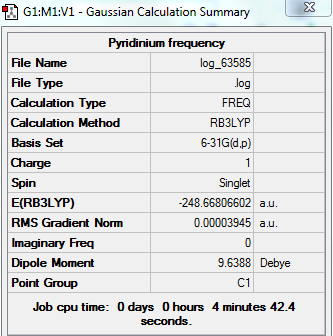
Pyridinium
Optimised Pyridinium
test molecule |
Summary Table
Real Output--Item Table
Item Value Threshold Converged?
Maximum Force 0.000065 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000834 0.001800 YES
RMS Displacement 0.000177 0.001200 YES
Predicted change in Energy=-7.095348D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,5) 1.3838 -DE/DX = 0.0 !
! R3 R(1,6) 1.0835 -DE/DX = 0.0 !
! R4 R(2,3) 1.3988 -DE/DX = 0.0 !
! R5 R(2,7) 1.0852 -DE/DX = 0.0 !
! R6 R(3,4) 1.3839 -DE/DX = 0.0 !
! R7 R(3,8) 1.0835 -DE/DX = 0.0 !
! R8 R(4,9) 1.0832 -DE/DX = 0.0 !
! R9 R(4,12) 1.3523 -DE/DX = 0.0001 !
! R10 R(5,11) 1.0832 -DE/DX = 0.0 !
! R11 R(5,12) 1.3524 -DE/DX = 0.0 !
! R12 R(10,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 119.0799 -DE/DX = 0.0 !
! A2 A(2,1,6) 121.4986 -DE/DX = -0.0001 !
! A3 A(5,1,6) 119.4214 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0614 -DE/DX = 0.0 !
! A5 A(1,2,7) 119.9688 -DE/DX = 0.0 !
! A6 A(3,2,7) 119.9697 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.077 -DE/DX = 0.0 !
! A8 A(2,3,8) 121.5031 -DE/DX = -0.0001 !
! A9 A(4,3,8) 119.4199 -DE/DX = 0.0001 !
! A10 A(3,4,9) 123.9283 -DE/DX = 0.0 !
! A11 A(3,4,12) 119.2355 -DE/DX = 0.0 !
! A12 A(9,4,12) 116.8362 -DE/DX = 0.0 !
! A13 A(1,5,11) 123.9339 -DE/DX = 0.0 !
! A14 A(1,5,12) 119.2331 -DE/DX = 0.0 !
! A15 A(11,5,12) 116.833 -DE/DX = 0.0 !
! A16 A(4,12,5) 123.313 -DE/DX = 0.0 !
! A17 A(4,12,10) 118.344 -DE/DX = 0.0 !
! A18 A(5,12,10) 118.343 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0005 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0024 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0016 -DE/DX = 0.0 !
! D4 D(6,1,2,7) -0.0014 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0005 -DE/DX = 0.0 !
! D6 D(2,1,5,12) 0.0021 -DE/DX = 0.0 !
! D7 D(6,1,5,11) -0.0014 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.0011 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0022 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0017 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -179.9993 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0012 -DE/DX = 0.0 !
! D13 D(2,3,4,9) -179.9998 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0013 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0003 -DE/DX = 0.0 !
! D16 D(8,3,4,12) 180.0008 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0014 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -180.0007 -DE/DX = 0.0 !
! D19 D(9,4,12,5) 180.0024 -DE/DX = 0.0 !
! D20 D(9,4,12,10) 0.0003 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.0031 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.001 -DE/DX = 0.0 !
! D23 D(11,5,12,4) -180.0008 -DE/DX = 0.0 !
! D24 D(11,5,12,10) 0.0014 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-Space Link
Frequency Analysis of Pyridinium
Summary Table
Low Frequency
Low frequencies --- -5.3602 0.0006 0.0006 0.0010 11.0206 14.1189 Low frequencies --- 391.5106 404.4654 620.4188
D-space link
MO Analysis of Pyridinium
D-Space link
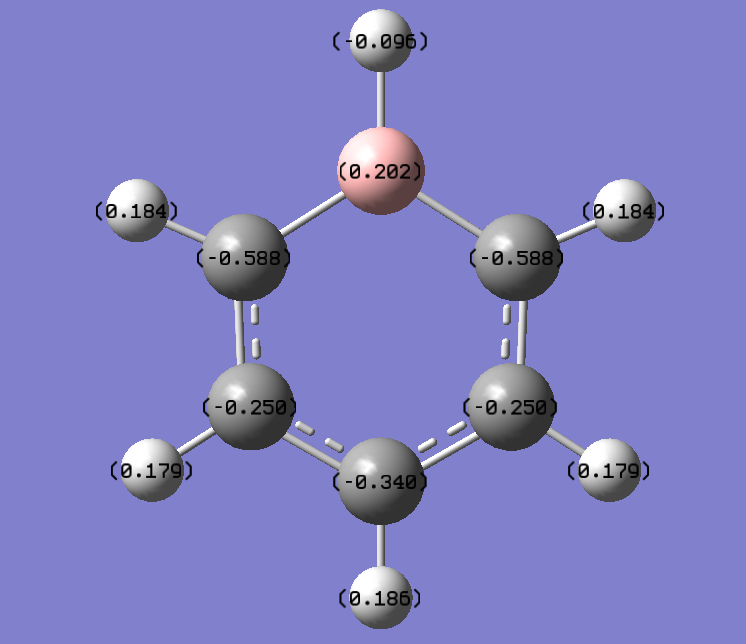
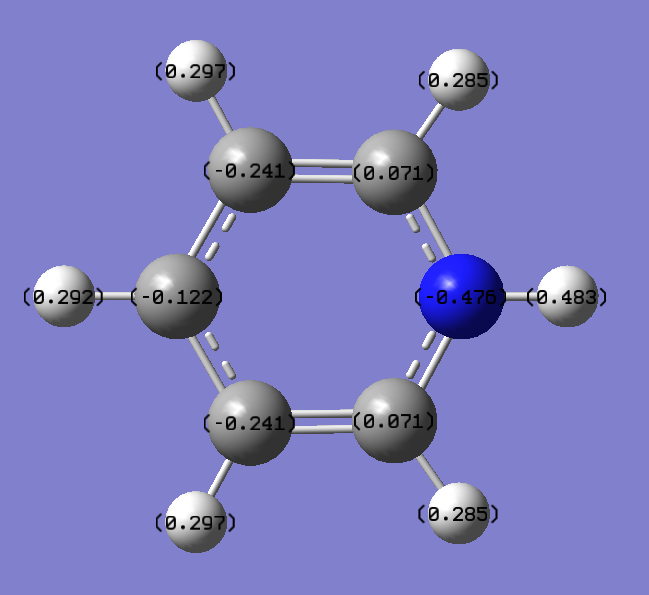
NBO Analysis for Pyridinium
Charge distribution
Charge Number
Real output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.24103 1.99912 4.22859 0.01331 6.24103
C 2 -0.12243 1.99913 4.10943 0.01386 6.12243
C 3 -0.24103 1.99912 4.22859 0.01331 6.24103
C 4 0.07101 1.99918 3.91066 0.01916 5.92899
C 5 0.07099 1.99918 3.91068 0.01916 5.92901
H 6 0.29719 0.00000 0.70178 0.00103 0.70281
H 7 0.29169 0.00000 0.70718 0.00113 0.70831
H 8 0.29719 0.00000 0.70178 0.00103 0.70281
H 9 0.28493 0.00000 0.71397 0.00110 0.71507
H 10 0.48278 0.00000 0.51476 0.00246 0.51722
H 11 0.28493 0.00000 0.71397 0.00110 0.71507
N 12 -0.47623 1.99937 5.46756 0.00929 7.47623
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98249) BD ( 1) C 1 - C 2
( 50.26%) 0.7089* C 1 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4187
-0.0371 -0.6896 0.0068 0.0003 0.0000
0.0122 0.0000 0.0000 -0.0118 -0.0115
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3938
-0.0234 0.7061 0.0290 -0.0003 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
2. (1.98297) BD ( 1) C 1 - C 5
( 49.58%) 0.7042* C 1 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 -0.0011 0.0320 -0.0006 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 5 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 0.0145 0.0331 0.0005 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
3. (1.61445) BD ( 2) C 1 - C 5
( 52.23%) 0.7227* C 1 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9997 -0.0068
0.0000 0.0191 -0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9995 -0.0175
0.0000 -0.0196 -0.0153 0.0000 0.0000
4. (1.97822) BD ( 1) C 1 - H 6
( 64.83%) 0.8052* C 1 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 -0.3985
0.0072 0.7230 -0.0181 0.0003 0.0000
-0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 6 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 0.0116 -0.0208 0.0000
5. (1.98249) BD ( 1) C 2 - C 3
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3931
-0.0235 -0.7065 -0.0289 -0.0003 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 3 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4181
-0.0371 0.6900 -0.0068 0.0003 0.0000
-0.0122 0.0000 0.0000 -0.0119 -0.0115
6. (1.54880) BD ( 2) C 2 - C 3
( 45.73%) 0.6762* C 2 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9997 -0.0036
0.0000 0.0241 -0.0101 0.0000 0.0000
( 54.27%) 0.7367* C 3 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9997 -0.0080
0.0000 0.0086 0.0228 0.0000 0.0000
7. (1.98141) BD ( 1) C 2 - H 7
( 64.64%) 0.8040* C 2 s( 31.07%)p 2.22( 68.90%)d 0.00( 0.03%)
0.0003 -0.5572 -0.0131 0.0007 0.8298
-0.0198 -0.0004 0.0000 -0.0006 0.0000
0.0000 0.0000 0.0000 -0.0153 0.0101
( 35.36%) 0.5947* H 7 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0242 0.0000 0.0000
8. (1.98297) BD ( 1) C 3 - C 4
( 49.58%) 0.7042* C 3 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 0.0004 -0.0320 -0.0006 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 4 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0047 -0.0138 -0.0331 0.0006 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
9. (1.97822) BD ( 1) C 3 - H 8
( 64.83%) 0.8052* C 3 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 0.3991
-0.0072 0.7226 -0.0181 -0.0003 0.0000
-0.0110 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 8 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 -0.0116 -0.0208 0.0000
10. (1.98154) BD ( 1) C 4 - H 9
( 64.26%) 0.8016* C 4 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 -0.4691
0.0193 0.6667 -0.0183 0.0004 0.0000
0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 9 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 0.0128 -0.0209 0.0000
11. (1.98861) BD ( 1) C 4 - N 12
( 36.68%) 0.6057* C 4 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5294 -0.0335 -0.0013 0.4046
0.0563 0.7415 0.0276 -0.0003 0.0000
0.0252 0.0000 0.0000 -0.0184 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 -0.3662
0.0186 -0.7067 -0.0132 0.0002 0.0000
0.0107 0.0000 0.0000 -0.0058 -0.0115
12. (1.82448) BD ( 2) C 4 - N 12
( 28.55%) 0.5343* C 4 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9991 0.0132
0.0000 0.0102 0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9999 0.0036
0.0000 -0.0128 -0.0077 0.0000 0.0000
13. (1.98154) BD ( 1) C 5 - H 11
( 64.26%) 0.8016* C 5 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 0.4697
-0.0193 0.6663 -0.0183 -0.0003 0.0000
0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 11 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0129 -0.0209 0.0000
14. (1.98861) BD ( 1) C 5 - N 12
( 36.68%) 0.6057* C 5 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4039
0.0563 -0.7418 -0.0277 -0.0003 0.0000
-0.0251 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 -0.3656
0.0187 0.7071 0.0132 0.0003 0.0000
-0.0107 0.0000 0.0000 -0.0059 -0.0115
15. (1.98629) BD ( 1) H 10 - N 12
( 25.41%) 0.5041* H 10 s( 99.88%)p 0.00( 0.12%)
0.9994 -0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.81%)p 2.73( 73.16%)d 0.00( 0.02%)
-0.0002 0.5178 0.0066 -0.0013 0.8553
-0.0091 -0.0004 0.0000 -0.0006 0.0000
0.0000 0.0000 0.0000 0.0115 -0.0106
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98249 -0.90378 118(v),114(v),107(g),63(v)
110(g),109(g),43(v),42(v)
112(g)
2. BD ( 1) C 1 - C 5 1.98297 -0.92650 120(v),112(v),106(g),33(v)
118(g),97(v),109(g),32(v)
119(g),96(v)
3. BD ( 2) C 1 - C 5 1.61445 -0.46666 111(v),117(v),99(v),108(g)
37(v)
4. BD ( 1) C 1 - H 6 1.97822 -0.71855 119(v),110(v),32(v),62(v)
106(g),107(g)
5. BD ( 1) C 2 - C 3 1.98249 -0.90379 115(v),109(v),113(g),54(v)
106(g),114(g),23(v),22(v)
112(g)
6. BD ( 2) C 2 - C 3 1.54880 -0.44892 117(v),108(v),56(v),26(v)
7. BD ( 1) C 2 - H 7 1.98141 -0.71804 107(v),113(v),22(v),42(v)
106(g),110(g)
8. BD ( 1) C 3 - C 4 1.98297 -0.92647 120(v),112(v),110(g),33(v)
115(g),97(v),114(g),32(v)
116(g),96(v)
9. BD ( 1) C 3 - H 8 1.97822 -0.71856 116(v),106(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - H 9 1.98154 -0.75116 119(v),110(v),42(v),113(g)
96(v)
11. BD ( 1) C 4 - N 12 1.98861 -1.06564 119(g),62(v),114(v),43(v)
118(v),63(v),113(g),120(g)
12. BD ( 2) C 4 - N 12 1.82448 -0.56812 108(v),111(v),64(v),46(v)
13. BD ( 1) C 5 - H 11 1.98154 -0.75116 116(v),106(v),22(v),107(g)
96(v)
14. BD ( 1) C 5 - N 12 1.98861 -1.06560 116(g),52(v),109(v),23(v)
115(v),54(v),107(g),120(g)
15. BD ( 1) H 10 - N 12 1.98629 -0.89228 113(v),107(v),52(v),62(v)
16. CR ( 1) C 1 1.99913 -10.26482 33(v),63(v),62(v),118(v)
110(v),72(v),112(v),119(v)
17. CR ( 1) C 2 1.99914 -10.27397 23(v),43(v),107(v),113(v)
114(v),109(v),76(v)
18. CR ( 1) C 3 1.99913 -10.26483 33(v),54(v),52(v),115(v)
106(v),80(v),112(v),116(v)
19. CR ( 1) C 4 1.99918 -10.32333 43(v),119(v),120(v),110(v)
113(g),97(v),84(v),114(v)
20. CR ( 1) C 5 1.99918 -10.32332 23(v),116(v),120(v),106(v)
107(g),97(v),92(v),109(v)
21. CR ( 1) N 12 1.99937 -14.46219 54(v),63(v)
Borazine
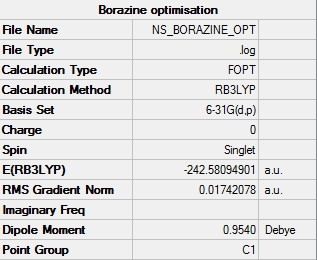
OPtimised Borazine
test molecule |
Summary Table
Real output--Item table
Item Value Threshold Converged?
Maximum Force 0.000117 0.000450 YES
RMS Force 0.000036 0.000300 YES
Maximum Displacement 0.000337 0.001800 YES
RMS Displacement 0.000104 0.001200 YES
Predicted change in Energy=-1.205717D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0097 -DE/DX = 0.0 !
! R2 R(2,12) 1.1949 -DE/DX = 0.0001 !
! R3 R(3,9) 1.0097 -DE/DX = 0.0 !
! R4 R(4,11) 1.1949 -DE/DX = 0.0001 !
! R5 R(5,7) 1.0097 -DE/DX = 0.0 !
! R6 R(6,10) 1.1949 -DE/DX = 0.0001 !
! R7 R(7,10) 1.4307 -DE/DX = 0.0 !
! R8 R(7,11) 1.4307 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4306 -DE/DX = 0.0 !
! R10 R(8,12) 1.4307 -DE/DX = -0.0001 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4306 -DE/DX = 0.0 !
! A1 A(5,7,10) 118.5554 -DE/DX = 0.0 !
! A2 A(5,7,11) 118.5605 -DE/DX = 0.0 !
! A3 A(10,7,11) 122.8841 -DE/DX = 0.0 !
! A4 A(1,8,10) 118.5623 -DE/DX = 0.0 !
! A5 A(1,8,12) 118.5671 -DE/DX = 0.0 !
! A6 A(10,8,12) 122.8706 -DE/DX = 0.0 !
! A7 A(3,9,11) 118.5663 -DE/DX = 0.0 !
! A8 A(3,9,12) 118.5599 -DE/DX = 0.0 !
! A9 A(11,9,12) 122.8739 -DE/DX = 0.0 !
! A10 A(6,10,7) 121.4339 -DE/DX = 0.0 !
! A11 A(6,10,8) 121.4439 -DE/DX = 0.0 !
! A12 A(7,10,8) 117.1222 -DE/DX = 0.0 !
! A13 A(4,11,7) 121.4457 -DE/DX = 0.0 !
! A14 A(4,11,9) 121.4368 -DE/DX = 0.0 !
! A15 A(7,11,9) 117.1175 -DE/DX = 0.0 !
! A16 A(2,12,8) 121.4314 -DE/DX = 0.0 !
! A17 A(2,12,9) 121.4369 -DE/DX = 0.0 !
! A18 A(8,12,9) 117.1317 -DE/DX = 0.0 !
! D1 D(5,7,10,6) -0.0006 -DE/DX = 0.0 !
! D2 D(5,7,10,8) 179.9989 -DE/DX = 0.0 !
! D3 D(11,7,10,6) 179.9979 -DE/DX = 0.0 !
! D4 D(11,7,10,8) -0.0026 -DE/DX = 0.0 !
! D5 D(5,7,11,4) 0.0008 -DE/DX = 0.0 !
! D6 D(5,7,11,9) 180.0001 -DE/DX = 0.0 !
! D7 D(10,7,11,4) -179.9977 -DE/DX = 0.0 !
! D8 D(10,7,11,9) 0.0015 -DE/DX = 0.0 !
! D9 D(1,8,10,6) 0.0011 -DE/DX = 0.0 !
! D10 D(1,8,10,7) -179.9984 -DE/DX = 0.0 !
! D11 D(12,8,10,6) -179.9993 -DE/DX = 0.0 !
! D12 D(12,8,10,7) 0.0012 -DE/DX = 0.0 !
! D13 D(1,8,12,2) -0.0012 -DE/DX = 0.0 !
! D14 D(1,8,12,9) 180.0007 -DE/DX = 0.0 !
! D15 D(10,8,12,2) -180.0008 -DE/DX = 0.0 !
! D16 D(10,8,12,9) 0.001 -DE/DX = 0.0 !
! D17 D(3,9,11,4) -0.0012 -DE/DX = 0.0 !
! D18 D(3,9,11,7) 179.9995 -DE/DX = 0.0 !
! D19 D(12,9,11,4) 180.0002 -DE/DX = 0.0 !
! D20 D(12,9,11,7) 0.001 -DE/DX = 0.0 !
! D21 D(3,9,12,2) 0.0011 -DE/DX = 0.0 !
! D22 D(3,9,12,8) -180.0008 -DE/DX = 0.0 !
! D23 D(11,9,12,2) 179.9997 -DE/DX = 0.0 !
! D24 D(11,9,12,8) -0.0022 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-space link
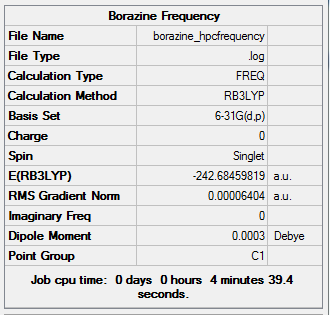
Frequency Analysis for Borazine
Summary Table
Low frequency
Low frequencies --- -10.7162 0.0005 0.0008 0.0009 7.9517 11.7161 Low frequencies --- 288.4992 290.4265 404.0112
D-space link
MO Analysis for Borazine
D-space Link
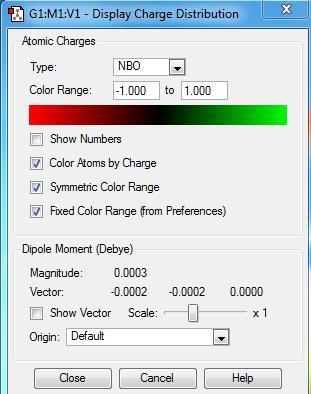
NBO Analysis for Borazine
Charge Distribution
Charge Number
Real output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 0.43199 0.00000 0.56573 0.00228 0.56801
H 2 -0.07655 0.00000 1.07586 0.00069 1.07655
H 3 0.43198 0.00000 0.56574 0.00228 0.56802
H 4 -0.07654 0.00000 1.07585 0.00069 1.07654
H 5 0.43197 0.00000 0.56575 0.00228 0.56803
H 6 -0.07655 0.00000 1.07586 0.00069 1.07655
N 7 -1.10239 1.99943 6.09818 0.00478 8.10239
N 8 -1.10239 1.99943 6.09818 0.00478 8.10239
N 9 -1.10239 1.99943 6.09818 0.00478 8.10239
B 10 0.74696 1.99917 2.23867 0.01520 4.25304
B 11 0.74697 1.99917 2.23865 0.01520 4.25303
B 12 0.74693 1.99917 2.23869 0.01520 4.25307
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06888 42.00000
NATURAL BOND ORBITAL ANALYSIS:
Occupancies Lewis Structure Low High
Occ. ------------------- ----------------- occ occ
Cycle Thresh. Lewis Non-Lewis CR BD 3C LP (L) (NL) Dev
=============================================================================
1(1) 1.90 40.69824 1.30176 6 12 0 3 3 3 0.03
2(2) 1.90 40.69824 1.30176 6 12 0 3 3 3 0.03
3(1) 1.80 41.27976 0.72024 6 15 0 0 0 3 0.03
-----------------------------------------------------------------------------
Structure accepted: No low occupancy Lewis orbitals
--------------------------------------------------------
Core 11.99579 ( 99.965% of 12)
Valence Lewis 29.28396 ( 97.613% of 30)
================== ============================
Total Lewis 41.27976 ( 98.285% of 42)
-----------------------------------------------------
Valence non-Lewis 0.67699 ( 1.612% of 42)
Rydberg non-Lewis 0.04325 ( 0.103% of 42)
================== ============================
Total non-Lewis 0.72024 ( 1.715% of 42)
--------------------------------------------------------
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98495) BD ( 1) H 1 - N 8
( 28.08%) 0.5299* H 1 s( 99.91%)p 0.00( 0.09%)
0.9996 -0.0010 -0.0252 -0.0154 0.0000
( 71.92%) 0.8481* N 8 s( 22.83%)p 3.38( 77.14%)d 0.00( 0.03%)
-0.0002 0.4776 -0.0114 0.0006 0.7498
0.0111 0.4572 0.0068 0.0000 0.0000
0.0108 0.0000 0.0000 0.0056 -0.0119
2. (1.98670) BD ( 1) H 2 - B 12
( 54.03%) 0.7351* H 2 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0005 -0.0192 0.0000
( 45.97%) 0.6780* B 12 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.0189
0.0006 0.7896 -0.0269 0.0000 0.0000
-0.0011 0.0000 0.0000 -0.0236 -0.0098
3. (1.98495) BD ( 1) H 3 - N 9
( 28.08%) 0.5299* H 3 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 -0.0259 0.0142 0.0000
( 71.92%) 0.8481* N 9 s( 22.83%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 0.7708
0.0115 -0.4209 -0.0063 0.0000 0.0000
0.0102 0.0000 0.0000 -0.0066 0.0119
4. (1.98670) BD ( 1) H 4 - B 11
( 54.03%) 0.7351* H 4 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0164 0.0100 0.0000
( 45.97%) 0.6780* B 11 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.6744
0.0230 -0.4112 0.0140 0.0000 0.0000
0.0210 0.0000 0.0000 0.0108 -0.0098
5. (1.98495) BD ( 1) H 5 - N 7
( 28.08%) 0.5299* H 5 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 0.0007 -0.0295 0.0000
( 71.92%) 0.8481* N 7 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 -0.0210
-0.0003 0.8780 0.0131 0.0000 0.0000
0.0006 0.0000 0.0000 0.0122 0.0119
6. (1.98670) BD ( 1) H 6 - B 10
( 54.03%) 0.7351* H 6 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 -0.0168 0.0092 0.0000
( 45.97%) 0.6780* B 10 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 0.6932
-0.0236 -0.3785 0.0129 0.0000 0.0000
-0.0199 0.0000 0.0000 0.0128 -0.0098
7. (1.98438) BD ( 1) N 7 - B 10
( 76.47%) 0.8745* N 7 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.6988
0.0003 0.3547 -0.0159 0.0000 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0085
( 23.53%) 0.4851* B 10 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.7164
-0.0200 -0.4104 -0.0543 0.0000 0.0000
0.0373 0.0000 0.0000 0.0256 -0.0206
8. (1.98438) BD ( 1) N 7 - B 11
( 76.47%) 0.8745* N 7 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7150
-0.0005 -0.3208 0.0159 0.0000 0.0000
0.0055 0.0000 0.0000 -0.0047 0.0085
( 23.53%) 0.4851* B 11 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.7352
-0.0225 0.3758 0.0532 0.0000 0.0000
0.0347 0.0000 0.0000 -0.0290 0.0206
9. (1.82090) BD ( 2) N 7 - B 11
( 88.21%) 0.9392* N 7 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 -0.0006 0.0046 0.0000 0.0000
( 11.79%) 0.3433* B 11 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 0.0578 -0.0206 0.0000 0.0000
10. (1.98438) BD ( 1) N 8 - B 10
( 76.47%) 0.8745* N 8 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.0796
-0.0135 0.7796 -0.0084 0.0000 0.0000
0.0013 0.0000 0.0000 0.0071 0.0085
( 23.53%) 0.4851* B 10 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.0421
-0.0348 -0.8246 -0.0462 0.0000 0.0000
0.0078 0.0000 0.0000 0.0445 0.0206
11. (1.82090) BD ( 2) N 8 - B 10
( 88.21%) 0.9392* N 8 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 -0.0037 -0.0028 0.0000 0.0000
( 11.79%) 0.3433* B 10 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 -0.0110 0.0604 0.0000 0.0000
12. (1.98438) BD ( 1) N 8 - B 12
( 76.47%) 0.8745* N 8 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 -0.6566
0.0136 0.4278 0.0082 0.0000 0.0000
-0.0065 0.0000 0.0000 0.0031 -0.0085
( 23.53%) 0.4851* B 12 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 0.7137
0.0570 -0.4151 0.0098 0.0000 0.0000
-0.0408 0.0000 0.0000 0.0195 -0.0206
13. (1.98438) BD ( 1) N 9 - B 11
( 76.47%) 0.8745* N 9 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.0423
0.0139 0.7825 -0.0077 0.0000 0.0000
-0.0006 0.0000 0.0000 0.0072 0.0085
( 23.53%) 0.4851* B 11 s( 31.25%)p 2.19( 68.51%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.0027
0.0370 -0.8257 -0.0444 0.0000 0.0000
-0.0035 0.0000 0.0000 0.0451 0.0206
14. (1.98438) BD ( 1) N 9 - B 12
( 76.47%) 0.8745* N 9 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.6354
-0.0140 0.4587 0.0075 0.0000 0.0000
0.0068 0.0000 0.0000 0.0024 -0.0085
( 23.53%) 0.4851* B 12 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5588 -0.0174 0.0032 -0.6930
-0.0574 -0.4488 0.0071 0.0000 0.0000
0.0425 0.0000 0.0000 0.0155 -0.0206
15. (1.82092) BD ( 2) N 9 - B 12
( 88.21%) 0.9392* N 9 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 0.0043 -0.0018 0.0000 0.0000
( 11.79%) 0.3433* B 12 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 -0.0468 -0.0398 0.0000 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H6B3N3)
1. BD ( 1) H 1 - N 8 1.98495 -0.61484 119(v),112(v),115(g),117(g)
76(v),96(v)
2. BD ( 1) H 2 - B 12 1.98670 -0.40393 118(v),115(v),66(v),56(v)
3. BD ( 1) H 3 - N 9 1.98495 -0.61484 117(v),113(v),119(g),118(g)
96(v),86(v)
4. BD ( 1) H 4 - B 11 1.98670 -0.40396 119(v),112(v),46(v),66(v)
5. BD ( 1) H 5 - N 7 1.98495 -0.61482 115(v),118(v),112(g),113(g)
76(v),86(v)
6. BD ( 1) H 6 - B 10 1.98670 -0.40394 113(v),117(v),46(v),56(v)
7. BD ( 1) N 7 - B 10 1.98438 -0.68871 113(g),106(v),110(g),109(v)
87(v),118(v)
8. BD ( 1) N 7 - B 11 1.98438 -0.68870 112(g),108(v),110(g),111(v)
77(v),115(v)
9. BD ( 2) N 7 - B 11 1.82090 -0.27139 116(v),82(v),78(v),39(v)
114(g)
10. BD ( 1) N 8 - B 10 1.98438 -0.68872 117(g),110(v),106(g),107(v)
97(v),119(v)
11. BD ( 2) N 8 - B 10 1.82090 -0.27139 120(v),102(v),98(v),23(v)
116(g)
12. BD ( 1) N 8 - B 12 1.98438 -0.68868 115(g),108(v),106(g),111(v)
77(v),112(v)
13. BD ( 1) N 9 - B 11 1.98438 -0.68869 119(g),110(v),108(g),107(v)
97(v),117(v)
14. BD ( 1) N 9 - B 12 1.98438 -0.68873 118(g),106(v),108(g),109(v)
87(v),113(v)
15. BD ( 2) N 9 - B 12 1.82092 -0.27140 114(v),92(v),88(v),31(v)
120(g)
16. CR ( 1) N 7 1.99943 -14.13096 77(v),87(v),112(g),113(g)
17. CR ( 1) N 8 1.99943 -14.13096 77(v),97(v),115(g),117(g)
18. CR ( 1) N 9 1.99943 -14.13096 87(v),97(v),119(g),118(g)
19. CR ( 1) B 10 1.99917 -6.65244 113(v),117(v),106(v),110(v)
20. CR ( 1) B 11 1.99917 -6.65246 112(v),119(v),108(v),110(v)
21. CR ( 1) B 12 1.99917 -6.65245 118(v),115(v),106(v),108(v)
Discussion
According to the graph above, the pi total bonding for these four molecules are similar. The difference is the electronegativity of the replacement element(N,B). The electronegativity of the N atom is greater than the carbon atom, then the electron is not sharing equally between two atoms . The delocalisation pattern is no more symmetric.
Charge number :
Benzene : C atom-> minus0.239 Hatom--> 0.239
Boratabenzene : B atom-->0.202 H atom-->0.184,0.179,-0.096, 0.186
Pyridinium: N atom-->-0.476 H atom-->0.285, 0.297. 0.483
Borazine: B atom-->-1.102 N atom-->0.747 H atoms-->-0.077, 0.432
question
Consider what effect the substitutions would have on the full MO diagram the LCAOs contributing to the MOs? the energy of the MOs? the degeneracy of the MOs?
Ans As the electronegativity of the atoms are not equal this time. So when we draw the LCAO, the atom with the large electronegativity is lying lower than the orther one which means contribute more to the MOs.