Victor's 2nd modelling
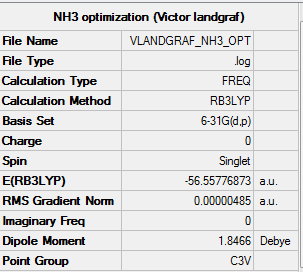
NH3 optimization
General information
H-N-H bond angle: 105.741°
N-H bond length: 101.798 pm
Charges: N has a higher electronegativity than H, thus one would expect a negative charge for N and a positive charge for H. Indeed calculations of GaussView yield the following charges: N: -1.125 ; H:0.375
Summary table
Item table with link to LOG file
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986258D-10 Optimization completed.
JSmol of optimised NH3 molecule
optimised NH3 molecule |
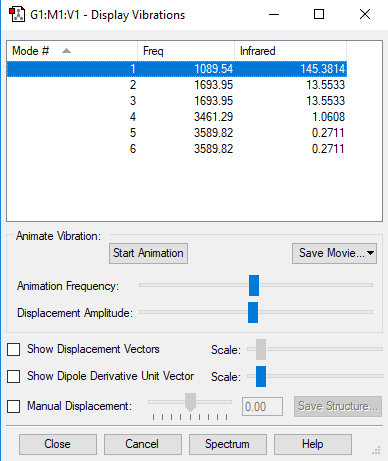
Vibrations
Answers to question
1. As there are four atoms in an NH3 molecule the 3N-6 rule predicts 6 modes.
2. Mode 2,3 and 5,6 have the same energy respectively: Their absorption frequencies are identical, thus they have to have the same energy as E=hf, where h is the Planck constant and f is the frequency.
3. 1,2,3 are bending modes, 4,5,and 6 are stretching modes. This can be deduced from the GaussView animation.
4. Mode 4 looks highly symmetric in the Gaussview animation.
5. The umbrella mode is mode 1 as can be seen from the animation on GaussView.
6. One would expect six absorptions but there are four as there are two pairs of identical absorption frequencies.
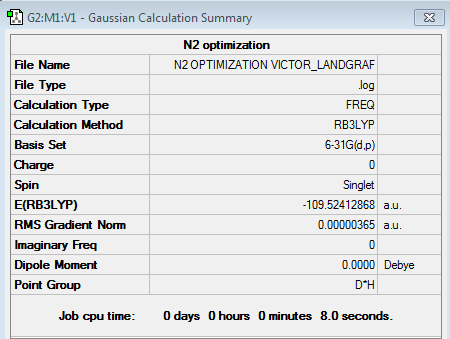
N2 optimisation
General information
Bond length of N2 : 110.5 pm
Bond angle: 180° as it is a linear molecule.
Summary table
item table with link to LOG file
File:N2 OPTIMIZATION VICTOR LANDGRAF.LOG
JSmol of optimised N2
opt. N2 molecule |
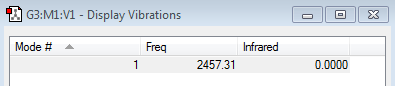
Vibrations
The 3N-5 rule predicts only one vibration as the N2 molecule contains 2 atoms. There is no dipole in N2 which is why it cannot be seen in IR spectroscopy (i.e the intesnity is 0).
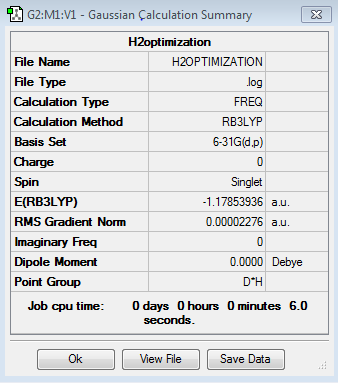
H2 optimisation
General information
Bond length of optimized molecule: 74.274 pm
Bond angle: 180° as it is a linear molecule.
Charges: Charges are the same as H2 contains two identical H atoms.
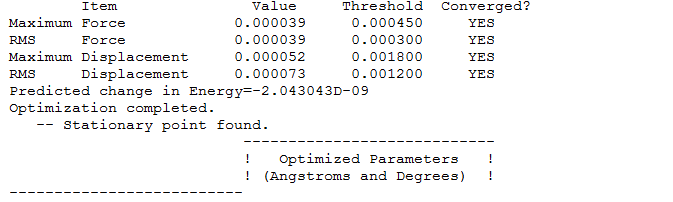
Summary table
Item table with LOG file
JSmol of optimised H2 molecule
opt. H2 molecule |
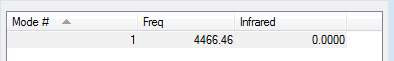
Vibrations
This consists with the expectation as the 3N-5 rule predicts 1 mode for the H2 molecule as it contains 2 atoms. There is no dipole in H2 which is why it cannot be seen in IR spectroscopy (i.e the intesnity is 0).
Haber-Bosch process
N2 + 3 H2 --> 2 NH3
E(NH3 )= -56.55776873 au
2*E(NH3 )= -113.1155375 au
E(N2 )= -109.52412868 au
E(H2 )= -1.17853936 au
3*E(H2 ) = -3.53561808 au
ΔE=2 E(NH3 )-[(E(N2 )+3 E(H2 )]
ΔE= -113.1155375 au - (-109.5241287-3.5356180)au = 0.0552280 au = -145.00 kJ/mol
The energy necessary to convert H2 gas and N2 gas into NH3 gas is -145.00 kJ/mol. This shows that the energy required to break the N2 and H2 bonds is less than the energy released forming the bonds that make NH3 . Thus the product is more stable.
ΔHf for the formation of 2 NH3 is -92 kJ/mol [1]
for 723K and 100 atm of pressure. The diverge because the conditions GaussView uses for its calculations are different and because the entropy changes are not taken into account properly by GaussView.
Reference
- ↑ Rob Davis Periodicity [Lecture] Imperial College, March 2016.
ClF my small molecule
General information
Bond length: 166.434 pm
Bond angle: 180° as it is a linear molecule.
Charges: -0.309 (F) and +0.309 (Cl) which is consistent with the expectations as F is more electronegative.
Summary table
Item table with link to LOG file
Link to LOG file: File:CLF OTPIMIZATION.LOG
JSmol of optimised ClF molecule
optimized ClF molecule |
Vibrations
ClF has 1 mode as the 3N-5 rule predicts (N=#of atoms in molecule)
Light absorption frequency: 781.00 Hz
5 Orbitals
Orbital1
Bonding sigma molecular orbital (MO) where one 3s of Cl and the 2s of F overlap. Energetically it is in the HOMO/LUMO region (-1.2186 au) compared to the HOMO -0.32855 au. It is occupied and would thus form a sigma bond between the two atoms but there is an occupied anti bonding MO (see below) which "cancels" this bond. It can be seen that the 2p atomic orbital (AO) of F contributes more to this bonding MO. This in the case because it is lower in energy than the 3p AO of Cl it interacts with.
Orbital2
Anti bonding sigma MO where 3s AO of Cl and the 2s AO of F overlap. It is energetically close to the HOMO/LUMO region, too. It is occupied and thus "cancels" the bond that would be formed by the occupied bonding MO above. It can be seen that the 3s AO of Cl contributes more to the anti bonding MO as it is higher in energy than the 2s AO of F.
Orbital3
Bonding sigma MO formed by overlap of 3p AO of Cl and 2p AO of F. It is in the HOMO/LUMO region, energy wise. The 2p AO of F contributes more to this bonding MO for reasons explained above. It is occupied and forms a bond between the two atoms. There is no occupied anti bonding orbital that would "cancel" this bond thus it consists.
Orbital4
Pi bonding orbital formed by overlap of 3p atomic orbital of Cl and 2p atomic orbital of F. It is in the HOMO/LUMO region, energy wise and occupied. This MO would form a bond but there is an occupied anti bonding MO that "cancels" the bond.
Orbital5
Pi anti bonding MO formed by overlap of 3p AO of Cl and 2p AO of F out of phase. Energetically it is in the HOMO/LUMO region. It is occupied and thus cancels the bond that would be formed by the occupied pi bonding molecular orbital above. It can be seen that the 3p AO of Cl contributes more to the anti bonding MO as it is higher in energy than the 2p AO of F.