Using the cluster : tutorial, examples
Connecting to the cluster
Running calculations on the cluster is quicker and you can submit big calculations more easily than you can on your Mac. You should be able to start using the cluster after you get your college username and password.
SSH
Secure Shell or SSH is a network protocol that allows data to be exchanged using a secure channel between two networked devices.
We use the ssh command to access the cluster, and build a connection like a safe tunnel between your mac and the cluster. Thus you can run and follow calculations on the cluster via your mac.
This can be used with a number of options (use the man ssh command to view these). One of the most useful is -Y which enables X11 forwarding, enabling graphical user interfaces (GUIs), such as nedit or xpbs, to be used.
How to use SSH protocol
To connect to the PC cluster you must have an IC college account.
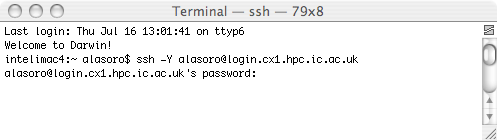
When you have an IC college account, open a Terminal shell for example:
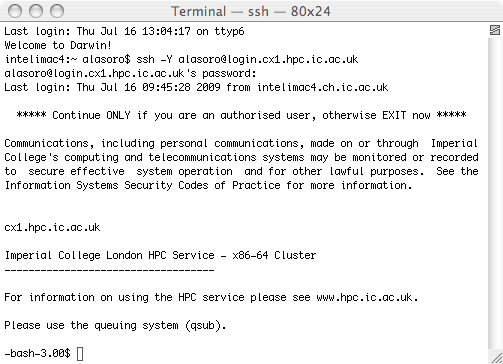
Then type ssh -Y username@login.cx1.hpc.ic.ac.uk. The username is the one given to you by Imperial College. When using this page, always substitute 'username' for your personal username.
It will ask you for your password. After entering you password and pressing enter, you will be connected to the cluster.
If you want to leave the cluster, just close the window.
You can connect many shells to the cluster to make different manipulations easier, and you can have Terminal and X11 connected to the cluster at the same time.
First steps on the cluster
Structure of the cluster
The cluster can be thought of as a hierarchy of directories like any UNIX system. On it your user area is divided into two sections, work and home. Input files are usually made and saved in home and output files are saved in work.
Basic UniX commands
The cluster uses UNIX commands in order to navigate through the file system. Below you will find some basic commands that you will use often. They may often be used with a number of options. Use man command-name or command-name help for information on these options. For further information the following two links are of interest:
UNIX Online Tutorials
List of UNIX commands
pwd
Displays current position in the file system.
-bash-3.00$ pwd /home/username -bash-3.00$
So, when you connect to the cluster you are in your HOME directory.
ls
Displays files and directories inside the directory you are in, which in the example here is '/home/username'.
-bash-3.00$ ls ONIOM2 benzene protein ONIOM bin qst2 g03 -bash-3.00$
In the /home/username directory there are different folders but if this is your first time on the cluster this is probably empty. We are going to create a folder in the next part.
mkdir
Creates a folder.
-bash-3.00$ mkdir tutorial -bash-3.00$
We now have a folder named tutorial in the 'username' directory. (The path to this folder is '/home/username/tutorial'). We can check that the folder exists with the ls command.
-bash-3.00$ ls ONIOM2 benzene protein ONIOM bin qst2 g03 tutorial -bash-3.00$
cd
This command takes you to a specific directory, and it allows you to change that directory. You must use the path-name of the folder you wish to move to.
-bash-3.00$ cd /work/username/ -bash-3.00$ pwd /work/username -bash-3.00$
Now you are in your WORK directory. Following the above steps a folder 'tutorial' can also be created here (within your username). Note that if you are already in your WORK directory, and want to change the 'username' directory in WORK, you can simply type 'cd username/'.
-bash-3.00$ pwd /work -bash-3.00$ cd username/ -bash-3.00$ pwd /work/username -bash-3.00$
cd ..
Move up a level out of the folder you are in.
-bash-3.00$ pwd /home/username/tutorial -bash-3.00$ cd .. -bash-3.00$ pwd -bash-3.00$ /home/username/
This is useful for navigating your way around directories.
Moving between the HOME and WORK directories
You have two directories in the cluster : a HOME folder and a WORK one. When you connect to the cluster you are directly in your HOME directory. In order to go to your WORK directory you can use the command cd /work/username/. You can also use the shortcut cd $WORK in order to directly get to your part of the WORK directory. If you want to go back to your HOME directory, use cd $HOME or cd ~.
jump
Use this command to go straight from where you are in the HOME directory to the equivalent point in the WORK directory (or vice versa). Note that for new users, jump is not included (it is a custom command, not a builtin function). It can be added by following the instructions in "Adding the jump command".
-bash-3.00$ pwd /home/username/tutorial -bash-3.00$ jump cd /work/username/tutorial -bash-3.00$ pwd /work/username/tutorial
Adding the jump command
If using jump results in the error "jump: command not found" then it can be added as a custom command. First, ensure that you have a bin directory where you can store custom commands. This should be in your HOME directory (/home/username/bin/) - if this does not exist use mkdir to create it. Next, inside bin create a new file called 'jump.sh'. See "Making and manipulating a new text file" for instructions. In this file, go into 'insert' mode and paste the following (using right click, paste):
#!/bin/bash
unset dname
unset jname
dname=$(pwd)
jname=$(echo $dname | cut -f 2 -d '/')
if [ "$jname" = "work" ] ; then
jname="${dname/work/home}"
echo "cd $jname"
else
jname="${dname/home/work}"
echo "cd $jname"
fi
cd $jname
Save and close this file. Next you need to create an alias (shortcut) to this command called jump. Custom aliases made by the user are typically saved in a file called '.bashrc' that exists in the HOME directory. Create this file if it does not exist. Inside this file, in a new line, type (or paste):
alias jump='source /home/username/bin/jump.sh'
Remember to change 'username' to your username and save the file. Now the alias is created, you need to tell the terminal to look for aliases by changing the '.bash_profile' file. Ensure '.bash_profile' exists and it has the line (create it if not):
source /home/username/.bashrc
This only needs to be done once, and will cause all aliases saved in '.bashrc' to be accessible whenever the terminal is opened.
Before moving on, try using all the commands above to make sure you are comfortable with them.
Making and manipulating a new text file
Hopefully you are now comfortable with moving around the directories. The next step is to make a new text file as a practise run for when you will be making input files for Gaussian.
Making a new file
First, make sure you are in the folder that you would like the file to be saved in. The text editor you will be using is called 'vim' or 'vi'. Type 'vi name_of_your_file.com', then press enter. The following screen will appear.
-bash-4.1$ vi name_of_your_file.com ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ ~ "name_of_your_file.com" [New File]
Vim has two modes: 'command' and 'insert'. When you create a new file, you are in command mode by default. To insert text you will need to change to insert mode by pressing 'i'. You will see the bottom of the screen now says -INSERT-. Type some text:
-bash-4.1$ vi name_of_your_file.com
My first vi file
~
~
~
~
~
~
~
-- INSERT --
To save your file, first go back into command mode by pressing the esc button, then type ':wq' and press enter.
Now you have made your first text file, practise the commands below in order to become confident with manipulating your file.
cp
Copy a file.
-bash-3.00$ pwd /home/username/tutorial -bash-3.00$ ls testjob_1.com testjob_2.com -bash-3.00$ cp testjob_2.com testjob_3.com -bash-3.00$ ls testjob_1.com testjob_2.com testjob_3.com -bash-3.00$
Here we have made a copy of testjob_2.com, the new file testjob_3.com.
rm
Remove a file.
-bash-3.00$ pwd /work/username -bash-3.00$ ls -bash-3.00$ tutorial -bash-3.00$ cd tutorial/ -bash-3.00$ ls testjob_1.com testjob_2.com -bash-3.00$ rm testjob_2.com -bash-3.00$ ls testjob_1.com -bash-3.00$
If a file is no longer needed it can be removed with this command. If you wish to remove a whole directory use rm-r before specifying its name to remove the folder and all the files inside. When you are in the cluster and are trying to remove a file, a line will appear saying 'rm: remove regular file 'name_of_file'?' with a prompt. Type 'y' and press enter to remove the file.
mv
Move a file to another folder.
-bash-3.00$ pwd /home/username/tutorial -bash-3.00$ ls testjob_1.com testjob_2.com testjob_3.com -bash-3.00$ mv testjob_3.com /home/username/ -bash-3.00$ ls testjob_1.com testjob_2.com testjob_3.com -bash-3.00$ cd .. -bash-3.00$ pwd /home/username -bash-3.00$ ls testjob_3.com tutorial -bash-3.00$
Preparing your first calculation
(This tutorial was put together when the default released version of Gaussian was Gaussian 03 / g03. It's now Gaussian 09 / g09, so use 'g09' with 'module load gaussian' in what's below. --Mjbear 11:56, 23 March 2011 (UTC))
Introduction
You most probably have performed some calculations on your Mac, so you know that in order to run a calculation you need an input file. But now the difference is that to compute this calculation you must have a jobscript as well as an input file. And as usual a calculation will create an output file, a .chk file and also a jobscript_ file in which you may find some important information if your calculation does not start.
Input
When making input files on your mac, at the very beginning of the file you may have put the 'Link 0 command', which contains information about the output file. This line always begins with a % symbol. When making an input file on the cluster, several Link 0 commands have to be included. First, specify how much memory you want the job to take up, on the next line state how many processors you want the job to use, and then specify the location of the output checkpoint file. In the example below, we want to use 1400MB of memory and one processor. We want the output files to go to the 'test' folder in the user's WORK directory.
%mem=1400MB %nproc=1 %chk=/work/ns4912/test/ethane.chk # opt hf/3-21g ethane optimisation 0 1 C1 C2 C1 1.5 H1 C2 1.0 C1 109.5 H6 C1 1.0 C2 109.5 H1 180.0 H4 C1 1.0 C2 109.5 H1 300.0 H5 C1 1.0 C2 109.5 H1 60.0 H2 C2 1.0 C1 109.5 H5 180.0 H3 C2 1.0 C1 109.5 H4 180.0
Open an input file
To open an input file you have made already, use the UNIX command vi
vi name_of_your_file.com
If you want to change your input file, go into insert mode, make the required changes and then go back into command mode and save the changes using ':wq' as described earlier. If you want to quit and you haven't made any changes, use ':q' instead. Finally, if you want to quit without saving the changes you have made, use ':q!'.
Jobscript
As mentioned before, to run a job on the cluster, you need a jobscript as well as an input file. This is simply a file that tells the cluster how to handle the job and where to put the output files. Input files - which take up relatively little space - are saved in the HOME directory. The log and checkpoint files which form the output are much larger, so they are sent to the WORK directory, which has more space. This is not done automatically - we need to tell the cluster to do this in the jobscript.
Making a jobscript
Jobscript files can be written in vim. The important difference is that they are saved with the extension '.sh', not '.com' like main input files. The jobscript for the ethane optimisation job above is shown here:
#PBS -l ncpus=1 #PBS -l mem=1500mb #PBS -l walltime=01:30:00 #PBS -j oe module load gaussian g09 < /home/ns4912/test/ethane.com > /work/ns4912/test/ethane.log
We have specified the number of processors, the memory, and the time we would like to allocate to the job. The line 'module load gaussian' tells the system to start up Gaussian. The last line gives the version of Gaussian to be used (g09), and then it gives the location of the input file (in the HOME directory) and where we want to put the output files (in the WORK directory).
Advice Break
How can I predict the time of a calculation?
That's a good question but there is no magical recipe! It depends on your the size of your system, the method and basis set that you are using (a larger basis set corresponds to a calculation that will take longer), and finally it depends on the calculation that you would like to compute (a single point energy calculation is quicker than an optimisation with the same system).
Submitting your calculation
After writing your input file and jobscript, you have to submit your job. To do this, open your input file, then if it is as you want it, save it and close it. Next, type the command 'qsub', followed by the full name of your jobscript file. Press enter.
-bash-4.1$ vi ethane.com -bash-4.1$ qsub job.sh 1478294.cx1b
Your job should now be running. The number following your command is the Job Id. To check the progress of your job (for example, to see whether it is already running or still queuing), use the 'qstat' command.
-bash-4.1$ qstat Job id Name User Time Use S Queue ---------------- ---------------- ---------------- -------- - ----- 1478294.cx1b job.sh ns4912 00:00:00 R medium -bash-4.1$ qstat Job id Name User Time Use S Queue ---------------- ---------------- ---------------- -------- - ----- 1478294.cx1b job.sh ns4912 00:00:00 R medium -bash-4.1$ qstat -bash-4.1$
The penultimate column tells us the status of the job. The 'R' in this case means the job is running. A 'Q' would mean the job is still queuing. You can use the 'qstat' command as many times as you like to check the status of your job. When using the command does not return information on the status of your job, but instead results in a blank prompt line, it means your job has finished running.
If your job is queueing for a long time, then the part of the cluster you need to use is overbooked. The part of the cluster your job is sent to depends on the time and memory that your job requires. Each part of the cluster has a limit to the amount of calculations it can run, which is why some calculations will end up queuing for a long time. In the example above, the job was running on an area of the cluster called 'medium'. To look at all the different parts and determine how full they are, use the qstat -q command.
-bash-3.00$ qstat -q
server: cx1
Queue Memory CPU Time Walltime Node Run Que Lm State
---------------- ------ -------- -------- ---- ----- ----- ---- -----
vlong 1900mb -- 72:00:00 -- 110 26 110 E R
submit -- -- -- -- 0 0 -- E R
short 1000mb -- 01:00:00 -- 7 76 25 E R
medium 1000mb -- 04:00:00 -- 5 0 25 E R
long 1900mb -- 22:00:00 -- 69 24 60 E R
q24 3900mb -- 48:00:00 -- 0 0 0 E S
q816 244gb -- 72:00:00 -- 18 14 17 E R
monster -- -- -- -- 0 0 0 E S
bench 122gb -- 72:00:00 -- 0 0 -- E S
fixed -- -- 72:00:00 -- 0 0 -- E R
q48 61gb -- 72:00:00 -- 137 195 120 E R
pqaerostr -- -- 300:00:0 -- 10 3 -- E R
ibtest -- -- 480:00:0 -- 0 0 60 D S
tng 16gb -- 48:00:00 -- 0 0 600 E S
heplt2 -- -- -- -- 0 0 -- E R
xdbg 61gb -- 00:15:00 -- 0 0 3 E R
pqese -- -- 150:00:0 -- 12 0 -- E R
pqph -- -- 120:00:0 -- 9 0 -- E R
pqfb -- -- 120:00:0 -- 1 0 -- E R
pqms -- -- 72:00:00 -- 32 3 -- E R
slong 1900mb -- 72:00:00 -- 0 0 -- E S
pqmb -- -- 72:00:00 -- 0 0 -- E R
pqjk -- -- 200:00:0 -- 2 0 -- E R
test8 -- -- -- -- 0 0 -- E R
pqtyc -- -- 72:00:00 -- 2 0 -- E R
pqmls -- -- 72:00:00 -- 0 0 -- E R
pqeph -- -- 999:00:0 -- 123 10 -- E R
pqnh -- -- 72:00:00 -- 0 0 -- E R
chemlab1 -- -- 72:00:00 -- 1 0 -- E R
pqrevans -- -- 650:00:0 -- 6 0 -- E R
pqneuro -- -- 96:00:00 -- 18 0 -- E R
pqchemeng -- -- 72:00:00 -- 18 4 -- E R
pqciveng -- -- 999:00:0 -- 1 0 -- E R
test82 -- -- 72:00:00 -- 6 1 -- E R
pqjc1 -- -- 650:00:0 -- 1 0 -- E R
p1mem 12gb -- 72:00:00 -- 16 41 -- E R
test81 -- -- 72:00:00 -- 6 8 -- E R
pqesebot -- -- 100:00:0 -- 31 3 -- E R
pqspat -- -- 72:00:00 -- 6 0 -- E R
testnh -- -- 72:00:00 -- 0 0 -- E R
pqastro -- -- 72:00:00 -- 2 0 -- E R
pqplee -- -- 1000:00: -- 20 1 -- E R
pqplasp -- -- -- -- 0 0 -- E R
pqtzaki -- -- 96:00:00 -- 9 0 -- E R
pqexss -- -- 72:00:00 -- 2 0 -- E R
pqdd -- -- 500:00:0 -- 1 0 -- E R
----- -----
681 409
-bash-3.00$
From the 'Run' and 'Que' columns, we can see how many jobs each part of the cluster is running, and how many jobs each part has queuing. If your job has been sent to an area that is very full, you can try changing the time or memory you have requested for the job. By doing this and resubmitting the job, you may end up sending it to an area of the cluster that is less busy.
Cancel a calculation
If you wish to cancel a calculation which is running or queuing, use the 'qdel' command followed by the Job Id.
-bash-3.00$ qstat Job id Name User Time Use S Queue ---------------- ---------------- ---------------- -------- - ----- 2745894.cx1 jobscript_couma alasoro 0 Q p1mem -bash-3.00$ qdel 2745894.cx1 -bash-3.00$ qstat -bash-3.00$
Your results
Output file
To have a look at the output file, go to your WORK directory, and then to the folder that you have sent the output files to. You can then open the log file with vim (using 'vi name_of_your_file.log').
[intellimac4:~/g03] aurelie% vi aurelie_pyridine_iefpcm_water.log
Alternatively, you can use another text editor, such as 'less' or 'more'. To use 'less', type 'less name_of_your_file.log'. This allows you to look at the log file one page at a time. Press the space bar to go one window forward and 'B' to go one window up.
[intellimac4:~/g03] aurelie% less aurelie_pyridine_iefpcm_water.log
To use 'more', type 'more name_of_your_file.log'. This will show you your whole log file, which you may prefer.
Checkpoint file
The checkpoint file contains all the information concerning your system and the calculation that you computed, so it contains more information than you can find in your output file, but it is a binary file. This means that if you want to make the file understandable you will have to format it. It is not always necessary to look at the checkpoint file to get the information you need as the log file will contain lots of important information already. If you do want to format your checkpoint file, type 'module load gaussian' and then use the command 'formchk'.
Jobscript
When a calculation is running output and checkpoint files are created. A special jobscript file is also created which corresponds to this particular calculation. If your calculation failed, you can try to open the jobscript to see if it can give you an explanation.
-bash-3.00$ vi jobscript_tutor.o2760944
Using GaussView in the cluster
You should be familiar with using GaussView on your Mac. GaussView can also be used in the cluster. To open the program, start up the X11 application and type 'xhost +', then press enter. In a shell where you are logged in to the cluster, type 'module load gaussian' followed by 'gview &'.
-bash-4.1$ module load gaussian -bash-4.1$ gview & [1] 9486
Viewing output files in GaussView
It may be that you have written an input file in vim on the cluster and submitted it to Gaussian (as described above), but want to view only the output on GaussView. To do this, start up GaussView and open the checkpoint file created as part of your job's output. By default the 'File Type' is set to 'Gaussian Input Files'; to view your checkpoint file, change this to 'Gaussian Checkpoint Files'.
Doing an entire job in GaussView
Just as on the Mac, you can also do an entire job from start to finish in GaussView. The input files are created in your HOME directory, so the output files will also be put there. You can move them to your WORK directory using the 'mv' command.
Connecting the cluster to your Mac
You may wish to copy a file that you have saved on the cluster to your Mac, or you may wish to copy a file on your Mac to the cluster.
Copy a file from your Mac to your account on the cluster
This is done in a shell where you are not connected to the cluster. Use the 'scp' command, followed by the path to the file on your Mac. In the same line, login to the cluster and specify where on the cluster you would like the copy to be sent to.
intelimac1:~/g09 username$ scp /Users/username/g09/oxygen.com/ ns4912@login.cx1.hpc.ic.ac.uk:/home/ns4912 ns4912@login.cx1.hpc.ic.ac.uk's password: oxygen.com 100% 203 0.2KB/s 00:00
In this example, there a file called 'oxygen.com', which is saved in a folder called g09 in the user's area on the Mac. It is going to be copied to the user's area in the HOME directory on the cluster. After typing your command, you are prompted for your password. After entering it, the file is copied to the cluster. This can be double-checked when you log into the cluster.
-bash-4.1$ pwd /home/ns4912 -bash-4.1$ ls bin oxygen.com test
The file 'oxygen.com' now also appears in the user's area in the HOME directory on the cluster.
Copy a file from your account on the cluster to your Mac
Again, this is done in a shell where you are not connected to the cluster. The 'scp' command is also used here, but the order of phrases in your command is slightly different. In the example below, we will copy a file called 'ethane.chk' from the user's area in the WORK directory on the cluster to their area on the Mac. We will then check that the file has been successfully copied.
intelimac1:~ nafisa$ cd /Users/username/ intelimac1:~ nafisa$ ls Desktop Library Music Public g09 scripts Documents Movies Pictures Sites h2o.rtf work intelimac1:~ nafisa$ scp ns4912@login.cx1.hpc.ic.ac.uk:/work/ns4912/test/ethane.chk/ /Users/username/ ns4912@login.cx1.hpc.ic.ac.uk's password: ethane.chk 100% 768KB 768.0KB/s 00:00 intelimac1:~ nafisa$ cd /Users/username/ intelimac1:~ nafisa$ ls Desktop Library Music Public ethane.chk h2o.rtf work Documents Movies Pictures Sites g09 scripts
The file has been successfully copied to the Mac.
More about UNIX
This website lists classic UNIX commands: Basis UNIX commands