User talk:Ny210
Module 2
BH3 molecule
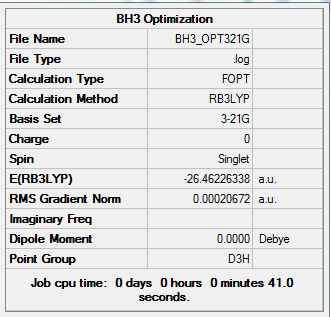
Analysing the optimised BH3 molecule using 3-21G basis set
For the optimization, the nuclei are assumed to be at given positions and the energy is evaluated by using the Schrodinger equation. Then the nuclei are moved and SCF cycle is repeated at various geometries. The program would compare and choose the geometry with the lowest energy. The created BH3 molecule with a trigonal planar structure is firstly optimised using B3LYP method and a low accuracy 3-21G basis set to give a rough idea of determining the optimum position of the nuclei.
Optimised BH3 molecule
BH molecule using a 3-21G basis set |
General Information
| B-H Bond Length | 1.19Å |
| H-B-H Bond Angle | 120.0° |
Initially the bond length of B-H is set to be 1.5 Å. After optimization, all the B-H lengths are observed to change to 1.19 Å, whereas the H-B-H angle is still 120° since the molecule belongs to the D3h point group. Also, the dipole moment is shown to be 0 Debye due to the symmetrical character of D3h point group.
Summary of Optimised BH3 molecule
After checking with the obtained summary and the output file, the molecule is found to be optimized successfully. This is because both the force and the gradient are converged since an energy minimum has been reached.
Real Output of Optimised BH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad


The plot of total energy against optimization step and the plot of RMS gradient norm against optimization step illustrate the decrease of the total energy and RMS gradient when the optimization calculation proceeds.
File Link
The optimisation file is liked to here
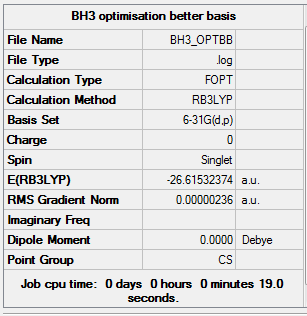
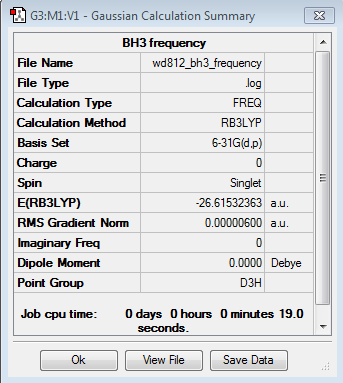
Analysing the optimised optimised BH3 molecule using 6-31G(d,p) basis set
A second optimization calculation for BH3 molecule is performed using the same B3LYP method but a higher level 6-31G(d,p) basis set with symmetry turning off in order to get rid of the influence of symmetry and reach the energy minimum correctly.
Optimised BH3 molecule
BH molecule using a 6-31G(d,p) basis set |
General Information
| B-H Bond Length | 1.19Å |
| H-B-H Bond Angle | 120.0° |
Summary of Optimised BH3 molecule
The point group is turned to Cs which confirms that the calculation switched off the symmetry.
Real Output of Optimised BH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000020 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.312911D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0002 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0002 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9997 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
File Link
The optimisation file is liked to here
Energy Differences in Using Different Basis Set
Total Energy
3-21G Basis Set: -26.46226338 a.u.
6-31G(d,p) Basis Set: -26.61532374 a.u.
The total energy for the calculation depends on the level of the basis set used. The energy difference in atomic units is observed to be small. However, when converting the units to kJ/mol as below,
3-21G Basis Set: 69476.6725kJ/mol
6-31G(d,p) Basis Set: 69878.53248 kJ/mol
The energy difference becomes 401.85998 kJ/mol which is quite large. And this confirms that comparison can only be made on the molecules with exactly the same basis set.
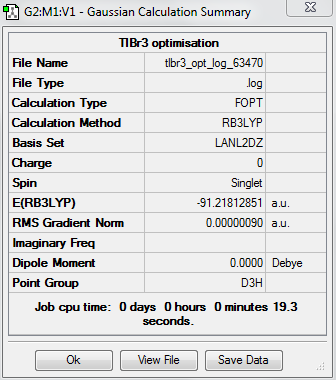
TlBr3 molecule
The optimization calculation for TlBr3 molecule is carried out using the B3LYP method but with a medium level LanL2DZ basis sets. It involves the use of pseudo potentials which can make the calculation easier and faster for molecules containing heavy atoms, such as thallium and bromine.
the Optimised TlBr3 Molecule Using Pseudo Potentials and LanL2DZ Basis Set
test molecule |
Summary of Optimised TlBr3 molecule
Real Output of Optimised TlBr3 molecule
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.083975D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The RMS gradient Norm is less than 0.00 and all the items are converged. This confirms that the optimization process is successful.
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20441
Comparison between optimised bond distance and literature data
| Optimization | Literature[1] | |
|---|---|---|
| Tl-Br Bond Length | 2.65Å | 2.52Å |
| Br-Tl-Br Bond Angle | 120.0° | 120° |
The obtained value defers a little from the literature value. This should be mainly because a medium level basis set is used or the optimisation is monitored in the gas phase. Therefore, the value may become more similar to the literature if we use basis sets with higher levels.
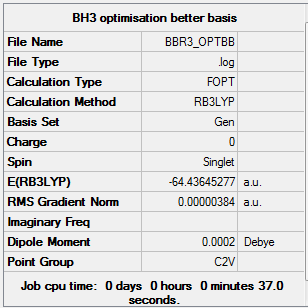
BBr3 molecule
Analysing the optimised BBr3 molecule using a mixture of basis set and pseudo potentials
A mixture of basis sets and pseudo potentials are used in the optimization of BBr3 molecule because the molecule involves both light B atom and heavy Br molecule. For the B atom, a 6-31G(d,p) basis set is used( from the previous BH3 file), whereas LanL2DZ basis sets are used on the Br atom.
Optimised BBr3 molecule
test molecule |
General Information
| B-Br Bond Length | 1.93Å |
| Br-B-Br Bond Angle | 120.0° |
Summary of Optimised BBr3 molecule
In the summary, the dipole moment becomes non-zero which may due to the 3 heavy electronegative Br atoms surrounding the small B atoms.
Real Output of Optimised BBr3 molecule
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.098477D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.9339 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0022 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0022 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9956 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The data above confirmed that the molecule is optimised.
File Link
The optimisation file is liked to here
Analysing the Results
Bond Length Comparison
| BH3 | 1.19349 |
| BBr3 | 1.93394 |
| TlBr3 | 2.65095 |
The equilibrium bond length in a molecule is the distance between the centres of the two bonded atoms. It can be partitioned into contributions from each atom of the bonded pair. So the bond length is dependent on both atoms involved in a bonded pair.[2]
In the case of BH3 and BBr3, changing the ligand gives different bond lengths. The bond length of BBr3 is longer because the Br atom is more electronegative than the H atom. Since Br atom has a greater electronegativity, the sharing of electrons is more unequal and more polar. More electron density around the B atom is pulled towards Br nucleus, so the bond length becomes longer. Besides, the orbital overlap is another important factor. The 1s and 2p orbitals of the B atom can overlap more efficient with the 1s orbitals of the H atom than with the 4s and 4p orbitals of the Br atom because the size of orbitals of B atom and H atom are similar. The bigger size of the Br orbitals only allows poor overlap with the B atom. Both H atom and Br atom share 1 electron with B atom to make a covalent bond between them. However, the electron donated from H atom is in the 1s orbital but the electron donated from Br atom is in the 4p orbital. Thus the overlap between them and B atom is quite different.
When comparing the bond lengths of BBr3 and TlBr3, changing the central atom makes the bond length different since the bond length of BBr3 is found to be shorter than that of TlBr3. B and Tl are both in Group 13, so they have same number of valence electrons. BBr3 has an incomplete octet and the electron deficiency can be partially removed by B-Br pi-bonding. In this case, the electron from Br atom can be donated to partially occupy the vacant p orbital on the B atom. Tl atom is much heavier and has more diffuse orbitals, thus TlBr3 is less stable and the bonds get weaker due to the poor overlap between Tl and Br.
When doing the computational analysis, the program considers the bond as a region with high electron density. Sometimes the Gaussview program does not draw bonds that would normally being existed. This is because gaussview draws bonds based on a distance criteria, so when a pre-defined distance is exceeded, the probability of finding electron density between two atoms falls to a value that the optimization calculation could not recognize it as a bond. However, this does not mean the bond is not present but only indicates the bond formed could not allowed the program to find a right geometry with minimum energy.
A bond is the attraction between atoms that results in the formation of chemical substances. In this case, the bond is covalent bond which involves the sharing of pairs of electron densities between two atoms. The bond length and bond length can be affected by several factors, such as electronegativity, the size of orbitals, and the effective overlap between orbitals on atoms.
Frequency Analysis
Frequency analysis is the second derivative of the potential energy surface and being used to confirm the optimized molecule has minimum structure. A positive value of the second derivative gives us minimum, whereas negative value indicates a transition state is present or the optimization failed.
Frequency Analysis for BH3
The frequency analysis is carried out using the B3LYP method with a 6-31G(d,p) basis set.
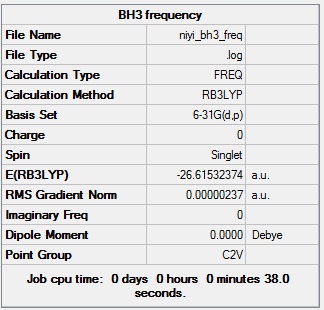
Summary of BH3 frequency
The total energy and the gradient is the same as the optimization calculation, which indicating the optimization is successful. However, the assigned point group changes from Cs to C2V.
Real Output of BH3 frequency
Low frequencies --- -18.6669 -0.0007 -0.0001 0.0008 12.5167 12.5631 Low frequencies --- 1162.9785 1213.1756 1213.2363
Every molecule has 3N-6 vibrational frequencies. The low frequencies listed above are the “-6”, and they are much smaller compared to the first vibration.
Animating the Vibrations of BH3
| no. | Type | Mode | Annotation | Frequency/cm-1 | Literature/cm-1[3] | Intensity | Symmetry D3h point group |
|---|---|---|---|---|---|---|---|
| 1 | Out of plane wagging |  |
Three H atoms move above and below the plane in a concerted manner, whereas the boron atom is wagging in the opposite direction. In this vibration, dipole moment changes a lot and thus the intensity is much stronger. | 1163 | 1225 | 93 | A2' |
| 2 | In plane scissoring |  |
Two H atoms scissor in a concerted manner, this results in the increase in two H-B-H bond angles and the decrease in the other one bond angle. There is only a small change in the dipole moment. | 1213 | 1305 | 14 | E' |
| 3 | In plane rocking |  |
Two H atoms rock in the plane whereas the other H atom rocks in the opposite direction. The change in the dipole moment is small. | 1213 | 1305 | 14 | E' |
| 4 | Symmetric stretching |  |
All H atoms stretch in and out in a concerted manner. The vibration is highly symmetrical, so the dipole moment cancels out. | 2582 | - | 0 | A1' |
| 5 | Asymmetric stretching |  |
Two H atoms stretch in the opposite direction, whereas the other H atom remains still. The change of dipole moment is quite large, so the intensity is strong. | 2715 | 2693 | 126 | E' |
| 6 | Asymmetric stretching |  |
Two H atom stretch in the same direction in a concerted manner, whereas the other one stretch in the opposite direction. There is also a large change in the dipole moment which results in the strong intensity. | 2712 | 2693 | 126 | E' |
IR spectrum of BH3
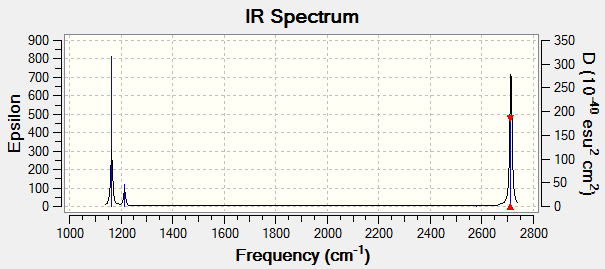
There should be 6 vibrational modes according to the 3N-6 rule, but the IR spectrum there are only three peaks. This could be explained by considering the vibrational frequencies and the intensities. The A1’ symmetry is totally symmetric, so it shows no intensity due to zero dipole moment change. Moreover, there are two sets of vibrations with symmetry E’ degenerate and thus each pair contributes to only one peak.
File Link
The frequency analysis file is liked to here
Frequency Analysis for TlBr3 molecule
The frequency analysis is calculated using the same basis set as the optimization.
Summary of TlBr3 Frequency
Real Output of TlBr3 frequency
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
Both the summary and the low frequencies term confirmed the optimization calculation is successful
Animating the Vibrations of TlBr3
| no. | Type | Mode | Annotation | Frequency/cm-1 | Literature/cm-1 [4] | Intensity | Symmetry D3h point group |
|---|---|---|---|---|---|---|---|
| 1 | In plane scissoring |  |
Two Br atoms scissor towards each other in the plane of the Tl atom. The other Br atom and the Tl atom move up and down in the opposite direction. The dipole moment changes a little, so it shows a weak intensity. | 46 | 47 | 4 | E' |
| 2 | In plane rocking |  |
Two Br atoms rock in the plane in a concerted manner, whereas the other Br rocks in the opposite direction. There is only a small change in the dipole moment. | 46 | 47 | 4 | E' |
| 3 | Out of plane wagging |  |
Three Br atoms move up and down the plane of Tl atom in a concerted manner, while the Tl atom is wagging in the opposite direction. The change in dipole moment is also small. | 52 | 63 | 6 | A2' |
| 4 | Symmetric stretching |  |
All Br atoms stretch in and out in a concerted manner. The vibration is highly symmetrical and there is no change in dipole moment. | 165 | - | 0 | A1' |
| 5 | Asymmetric stretching |  |
Two Br atoms stretch in the opposite direction, whereas the remaining Br atom remains stationary. There is a large change in dipole moment. | 211 | 203 | 25 | E' |
| 6 | Asymmetric stretching |  |
Two Br atoms stretch in the same direction in a concerted manner, while the other Br atom stretches in the opposite direction. There is also a large change in dipole moment, so the intensity is strong. | 211 | 203 | 25 | E' |
IR spectrum of TlBr3
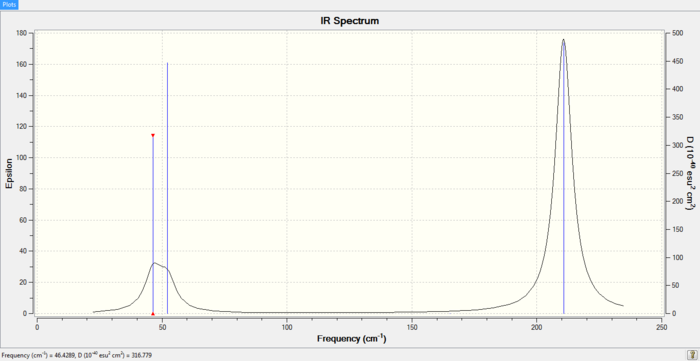
The A1’ symmetry could not be displayed because it showed no intensity. Again, two sets of vibrations with symmetry E’ are degenerate and thus each pair will only contribute to one peak.
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20484
Comparison of the Vibrational Frequencies of BH3 and TlBr3
| No. | Frequency of BH3 | Frequency of TlBr3 | Symmetry of BH3 | Symmetry of TlBr3 |
|---|---|---|---|---|
| 1 | 1163 | 46 | A2' | E' |
| 2 | 1213 | 46 | E' | E' |
| 3 | 1213 | 52 | E' | A2' |
| 4 | 2582 | 165 | A1' | A1' |
| 5 | 2715 | 211 | E' | E' |
| 6 | 2715 | 211 | E' | E' |
The same method and basis set is used because the total energy for any calculation depends highly on the level of basis set. The comparison should be based on the same energy level, or the result is not accurate.
From the data above, a large difference in the frequencies between the BH3 and TlBr3 molecules can be observed, and it is mainly due to the large bond strength difference and mass difference between the two molecules according to the equation below, v=1/2πc √(k/μ) , where μ is the reduced mass and k is the force constant. [5]
From the bond lengths obtained from previous investigation, bond length of Tl-Br is larger than that of B-H, which means Tl-Br bond is much weaker and thus gives lower vibrational frequencies. Moreover, the TlBr3 molecule has a much larger reduce mass which allows the molecule to have lower vibrational frequencies.
The two molecules have the same number of vibration modes and they show the same number of peaks in the IR spectra.
However there has been a reordering of modes for the two molecules since the A2” symmetry of BH3 molecule has the lowest vibrational frequencies whereas in TlBr3 one of the degenerate E’ symmetry pair has the lowest vibrational frequencies.
The two vibrational modes are close together due to the similarity in energy.
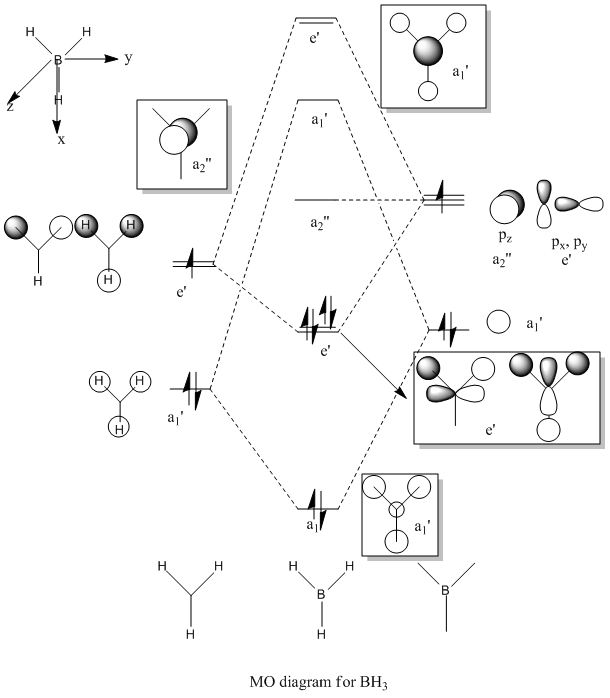
Molecular Orbitals of BH3
Completed Molecular Orbital
For predicting the MO, the energy calculation is carried out using the B3LYP method and a 6-31G(d,p) basis set, with the full population analysis and full NBO included.
When using VSEPR model, we could obtain the point group of the BH3 molecule as D3H. The fragments being chosen are H3 fragment and the B atom. The B atom is slightly more electropositive than the H3 fragment, thus the atomic orbitals of B atom need to be placed slightly higher in energy. The anti-bonding orbitals of the H3 fragment are only slightly higher in energy than the B 2s atomic orbital due to the small destabilizing interactions between H orbitals. Moreover, the 1s orbital of the B atom is not involved since it is too low in energy.
The shape of calculated molecular orbitals match up with the orbitals obtained from the LCAO approach. The computed molecular orbitals make it more easier to visualise how orbitals look like in real since LCAO approach could not provide continuous molecular orbitals as a whole. It can also express the distribution of electron density in a molecule.
However, 8 electrons are involved in the calculation other than 6 electrons in the LCAO approach since the low energy B 1s orbital is involved. Whether the s-s interactions is stronger than the s-p interactions is hard to rationalise in reality, but the 3a1' orbital is confirmed to be higher in energy than the 2e' orbitals in the calculation. Thus qualitative MO theory is useful for visualising the orbitals and ordering the energy for small molecules since the variations are small for quantum mechanical calculations. As the size of molecules increase, the accuracy becomes lower due to larger variations.
D space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20485
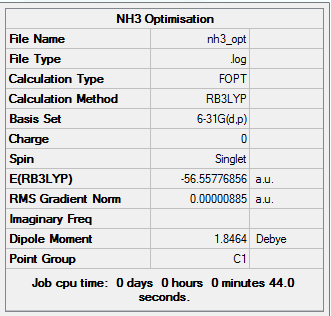
NH3 molecule
The NH3 molecule was optimized using the B3LYP method with a 6-31G(d,p) basis set because it does not involve any heavy atoms. The symmetry was turned off to get rid of the influence of the symmetry in giving the right energy.
Optimised NH3 molecule
test molecule |
General Information
| N-H Bond Length | 1.02Å |
| H-N-H Bond Angle | 105.7° |
Summary of Optimised NH3 molecule
Real Output of Optimised NH3 molecule
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629729D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
File Link
The optimisation file is liked to here
Frequency Analysis for NH3 molecule
Then it is analysed by frequency calculations using the previous optimized file.
Summary of NH3 Frequency
Real Output of NH3 frequency
Low frequencies --- -30.7178 -0.0012 -0.0011 0.0007 20.2209 28.2838 Low frequencies --- 1089.5549 1694.1246 1694.1858
Since the summary of both optimisation calculation and frequency calculation give similar results, the obtained geometry with minimum energy is confirmed.
IR spectrum of NH3
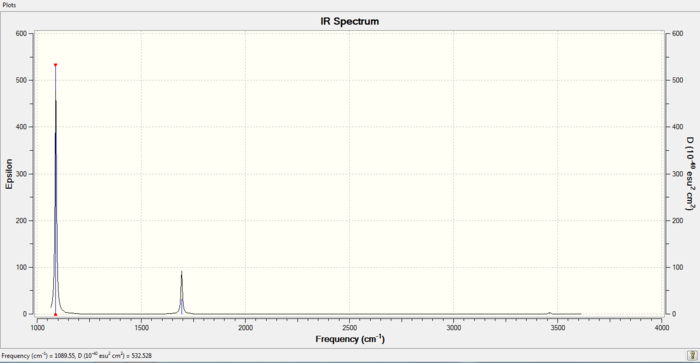
File Link
The frequency analysis file is liked to here
MO Analysis for NH3
The MO calculation of NH3 is carried out on the HPC service using B3LYP method with a 6-31G(d,p) basis set, involving a full population analysis and full NBOs.
File Link
The MO analysis file is liked to here
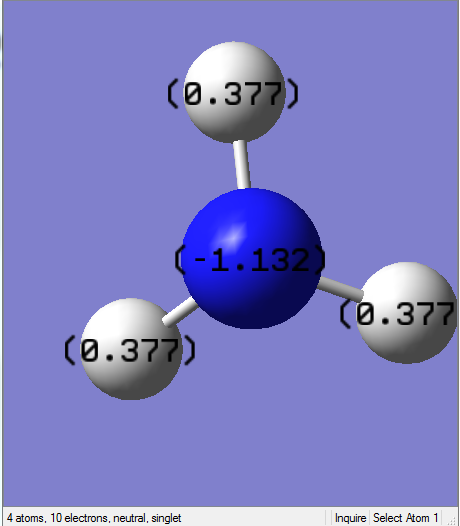
NBO Analysis for NH3
NBO analysis is carried on using the data from the previous population analysis. The charge limits for NBO analysis are set to be fixed between -1.000 and 1.000. The green colour indicates positive region, whereas the red colour indicates negative region.
Charge Distribution of NH3
Specific NBO Charges
From the graphs, N atom has much higher charge density than the H atoms which can be explained by the electronegativity difference. Since the N atom is more electronegative, it pulls electron density from the surrounding H atoms which gives rise to high electron density around it.
Real Output of NBO Analysis for NH3
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.13220 1.99981 6.11805 0.01434 8.13220
H 2 0.37740 0.00000 0.62045 0.00214 0.62260
H 3 0.37740 0.00000 0.62045 0.00214 0.62260
H 4 0.37740 0.00000 0.62045 0.00214 0.62260
=======================================================================
* Total * 0.00000 1.99981 7.97941 0.02078 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99925) BD ( 1) N 1 - H 2
( 68.94%) 0.8303* N 1 s( 27.14%)p 2.68( 72.79%)d 0.00( 0.07%)
-0.0001 -0.5209 -0.0046 0.0000 -0.2489
0.0021 0.8155 0.0294 0.0000 0.0000
0.0250 0.0000 0.0000 0.0066 0.0071
( 31.06%) 0.5573* H 2 s( 99.92%)p 0.00( 0.08%)
-0.9996 -0.0001 0.0056 -0.0285 0.0000
2. (1.99925) BD ( 1) N 1 - H 3
( 68.94%) 0.8303* N 1 s( 27.14%)p 2.68( 72.79%)d 0.00( 0.07%)
0.0001 0.5209 0.0046 0.0000 0.2489
-0.0021 0.4078 0.0147 0.7063 0.0254
0.0125 0.0216 0.0082 0.0005 0.0051
( 31.06%) 0.5573* H 3 s( 99.92%)p 0.00( 0.08%)
0.9996 0.0001 -0.0056 -0.0143 -0.0247
3. (1.99925) BD ( 1) N 1 - H 4
( 68.94%) 0.8303* N 1 s( 27.14%)p 2.68( 72.79%)d 0.00( 0.07%)
0.0001 0.5209 0.0046 0.0000 0.2489
-0.0021 0.4078 0.0147 -0.7063 -0.0254
0.0125 -0.0216 -0.0082 0.0005 0.0051
( 31.06%) 0.5573* H 4 s( 99.92%)p 0.00( 0.08%)
0.9996 0.0001 -0.0056 -0.0143 0.0247
4. (1.99981) CR ( 1) N 1 s(100.00%)
1.0000 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99756) LP ( 1) N 1 s( 18.55%)p 4.39( 81.37%)d 0.00( 0.08%)
0.0001 0.4305 -0.0115 0.0000 -0.9004
0.0539 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0246 0.0142
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99925 -0.61647
2. BD ( 1) N 1 - H 3 1.99925 -0.61648
3. BD ( 1) N 1 - H 4 1.99925 -0.61648
4. CR ( 1) N 1 1.99981 -14.15847
5. LP ( 1) N 1 1.99756 -0.28730 16(v),20(v),24(v),17(v)
21(v),25(v)
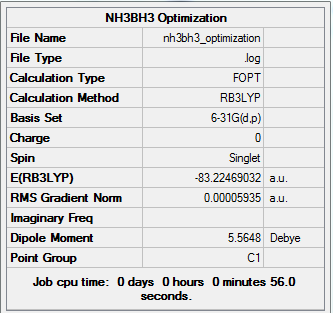
Ammonia-Borane Molecule
Due to its high hydrogen content and the character of acid-base complex, ammonia borane is being interested. The energy of the dative bond is going to be investigated as below.
Optimised Ammonia-Borane Molecule
The ammonia-borane molecule is optimised without symmetry using B3LYP method with 6-31G(d,p) basis set, this is because same level of basis sets need to be used in analysing the product molecule.
test molecule |
General Information
| B-H Bond Length | 1.21Å |
| H-B-H Bond Angle | 113.9° |
| N-H Bond Length | 1.02Å |
| H-N-H Bond Angle | 107.9° |
Summary of Optimised Ammonia-Borane Molecule
Real Output of Optimised Ammonia-Borane Molecule
Item Value Threshold Converged?
Maximum Force 0.000121 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000508 0.001800 YES
RMS Displacement 0.000294 0.001200 YES
Predicted change in Energy=-1.611643D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,8) 1.21 -DE/DX = -0.0001 !
! R5 R(5,8) 1.21 -DE/DX = -0.0001 !
! R6 R(6,8) 1.21 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.873 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.873 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0212 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.8692 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0298 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0297 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.8796 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8796 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.5972 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.8728 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.591 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.591 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9998 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9967 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.9963 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0005 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.003 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 179.996 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0001 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9964 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.0034 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The gradient is very small and the optimisation is converged.
File Link
The optimisation file is liked to here
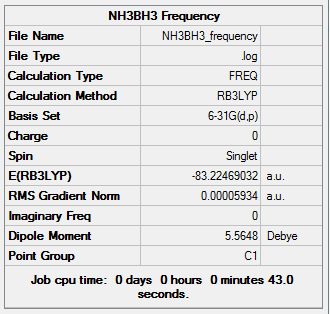
Frequency Analysis for Ammonia-Borane Molecule
The frequency analysis is carried out using the previous optimisation file.
Summary of Ammonia-Borane Frequency
Real Output of Ammonia-Borane frequency
Low frequencies --- -0.0014 -0.0002 0.0003 18.5341 23.7742 41.0264 Low frequencies --- 266.2856 632.2308 639.8248
From the frequency analysis above, we can ensure that we have a minimum since the low frequencies are low and negative frequencies are not present.
IR spectrum of Ammonia-Borane Molecule
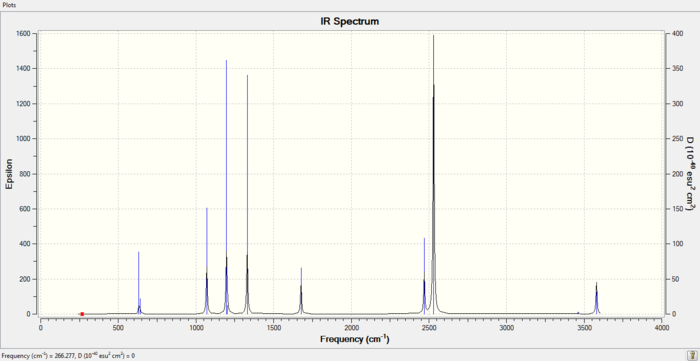
File Link
The frequency analysis file is liked to here
Analysing the Association Energies of Ammonia-Borane Molecule
E(NH3)=-56.55776856 a.u.
E(BH3)=-26.61532374 a.u.
E(NH3BH3)=-83.22469032 a.u.
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
=(-83.22469032)-[(-56.55776856)+(-26.61532374)]
=(-83.22469032)-(-83.1730923)
= 0.05159806 a.u.
= -135.4706015 kJ/mol
The value is sensible for a bond dissociation energy although the bond is weak in this case.
Accuracy
The energy would have an error of approximately 10 kJ/mol.
By converting it into a.u., the error would be
10 / 2625.50 = 0.0038088 a.u.
The energy depends highly on the method and the basis set used, thus a result with higher accuracy could be obtained by using a higher level basis set or a different computation method.
Mini Project
Benzene molecule is known as an aromatic compound, this project is aiming to investigate and compare its isoelectronic compounds by analysing the geometry, vibrations, molecular orbitals and NBOs.
Benzene Molecule
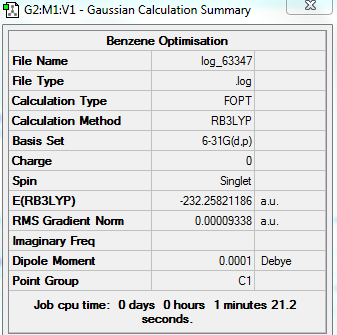
Optimised Benzene Molecule
The optimization calculation of benzene molecule is carried out using a BL3YP method with a 6-31G(d,p) basis set. The symmetry is being switched off.
test molecule |
General Information
| C-C Bond Length | 1.40Å |
| C-C-C Bond Angle | 120.0° |
Through the optimisation, all C-C bond lengths are equal to a value greater than a double bond but shorter than a single bond which gives rise to electron delocalization. The benzene molecule is planar and all the bond angles are equal to 120.0°.
Summary of Optimised Benzene Molecule
Real Output of Optimised Benzene Molecule
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000084 0.000300 YES
Maximum Displacement 0.000870 0.001800 YES
RMS Displacement 0.000313 0.001200 YES
Predicted change in Energy=-4.983462D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.0861 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.0861 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9996 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9968 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0036 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0036 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9916 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0048 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9967 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0101 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9932 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9996 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9891 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0113 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.004 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0048 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9912 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9965 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0072 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9963 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0021 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.0099 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0016 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.0068 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0119 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0087 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0039 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0007 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0064 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0058 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9968 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0026 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.005 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0061 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0057 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0055 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0085 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both the displacement and the force are converged, which confirms the geometry with a minimum.
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20594
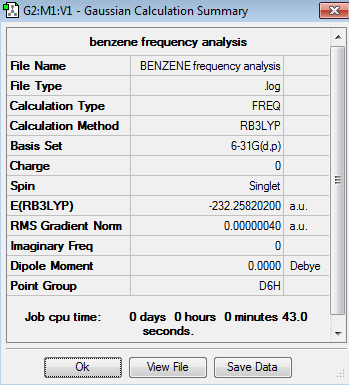
Frequency Analysis for Benzene Molecule
The frequency analysis is carried out using the previous optimisation file.
Summary of Benzene Frequency
Real Output of Benzene frequency
Low frequencies --- -14.2197 -2.6939 -0.0002 0.0005 0.0005 10.0128 Low frequencies --- 413.7276 414.5537 621.0443
The energy of the frequency calculation equals to that of the optimization calculation, and the low frequencies confirmed that a minimum energy state has been reached by the calculation.
IR spectrum of Benzene Molecule
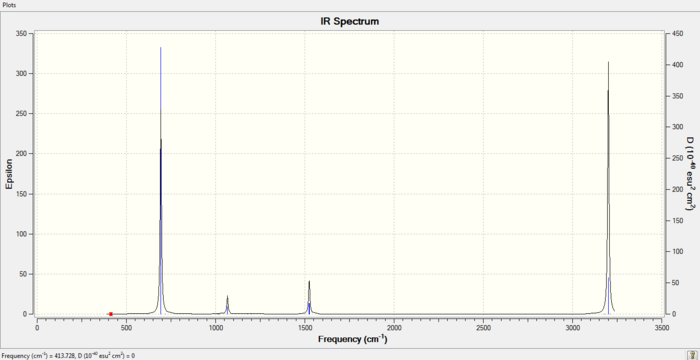
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20595
NBO Analysis for Benzene Molecule
The MO and NBO analysis is completed using the previous file, involving a full population analysis and full NBOs. The charge range is fixed between -1.000 and 1.000, and how charge distributes is shown below.
Charge Distribution of Benzene molecule
Specific NBO Charges
Real Output of NBO Analysis for Benzene Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.23856 1.99910 4.22615 0.01331 6.23856
C 2 -0.23855 1.99910 4.22613 0.01331 6.23855
C 3 -0.23853 1.99910 4.22612 0.01331 6.23853
C 4 -0.23856 1.99910 4.22615 0.01331 6.23856
C 5 -0.23854 1.99910 4.22613 0.01331 6.23854
C 6 -0.23853 1.99910 4.22612 0.01331 6.23853
H 7 0.23855 0.00000 0.76001 0.00144 0.76145
H 8 0.23854 0.00000 0.76002 0.00144 0.76146
H 9 0.23855 0.00000 0.76002 0.00144 0.76145
H 10 0.23855 0.00000 0.76001 0.00144 0.76145
H 11 0.23854 0.00000 0.76002 0.00144 0.76146
H 12 0.23855 0.00000 0.76002 0.00144 0.76145
=======================================================================
* Total * 0.00000 11.99462 29.91692 0.08846 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98096) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.7709
-0.0075 -0.2281 -0.0349 -0.0003 0.0000
0.0069 0.0000 0.0000 0.0151 -0.0109
( 50.00%) 0.7071* C 2 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.7816
0.0240 0.1884 -0.0264 0.0002 0.0000
0.0096 0.0000 0.0000 0.0135 -0.0109
2. (1.98097) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.2276
0.0349 0.7710 0.0076 0.0006 0.0000
0.0069 0.0000 0.0000 -0.0151 -0.0109
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.1878
0.0264 -0.7817 -0.0240 -0.0006 0.0000
0.0096 0.0000 0.0000 -0.0135 -0.0109
3. (1.66532) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0052 0.0188 0.0000 0.0000
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0139 -0.0137 0.0000 0.0000
4. (1.98305) BD ( 1) C 1 - H 7
( 62.04%) 0.7876* C 1 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.5933
-0.0103 -0.5930 0.0103 -0.0003 0.0000
-0.0166 0.0000 0.0000 0.0000 -0.0105
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0161 0.0161 0.0000
5. (1.98097) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.5830
-0.0340 0.5535 -0.0109 0.0004 0.0000
-0.0165 0.0000 0.0000 -0.0015 -0.0109
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.5539
-0.0109 -0.5826 -0.0340 -0.0004 0.0000
-0.0165 0.0000 0.0000 0.0015 -0.0109
6. (1.66532) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0049 0.0189 0.0000 0.0000
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 0.0189 -0.0049 0.0000 0.0000
7. (1.98305) BD ( 1) C 2 - H 8
( 62.04%) 0.7876* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.2167
-0.0038 0.8104 -0.0141 0.0006 0.0000
-0.0083 0.0000 0.0000 0.0144 0.0105
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0059 -0.0220 0.0000
8. (1.98096) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.1879
-0.0264 0.7817 0.0240 0.0006 0.0000
0.0096 0.0000 0.0000 -0.0135 -0.0109
( 50.00%) 0.7071* C 4 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.2276
-0.0349 -0.7711 -0.0076 -0.0005 0.0000
0.0069 0.0000 0.0000 -0.0151 -0.0109
9. (1.98305) BD ( 1) C 3 - H 9
( 62.04%) 0.7876* C 3 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.8102
-0.0141 0.2173 -0.0038 0.0002 0.0000
-0.0083 0.0000 0.0000 -0.0144 0.0105
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0220 -0.0059 0.0000
10. (1.98097) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.7709
0.0076 0.2281 0.0349 0.0002 0.0000
0.0069 0.0000 0.0000 0.0151 -0.0109
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.7815
-0.0240 -0.1884 0.0264 -0.0003 0.0000
0.0096 0.0000 0.0000 0.0135 -0.0109
11. (1.66534) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 0.0188 -0.0052 0.0000 0.0000
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 -0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0137 -0.0139 0.0000 0.0000
12. (1.98305) BD ( 1) C 4 - H 10
( 62.04%) 0.7876* C 4 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.5934
-0.0103 -0.5929 0.0103 -0.0003 0.0000
0.0166 0.0000 0.0000 0.0000 0.0105
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0161 0.0161 0.0000
13. (1.98096) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.5831
0.0340 -0.5536 0.0109 -0.0003 0.0000
-0.0165 0.0000 0.0000 -0.0015 -0.0109
( 50.00%) 0.7071* C 6 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.5540
0.0109 0.5826 0.0340 0.0003 0.0000
-0.0165 0.0000 0.0000 0.0015 -0.0109
14. (1.98305) BD ( 1) C 5 - H 11
( 62.04%) 0.7876* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.2169
-0.0038 0.8103 -0.0141 0.0006 0.0000
0.0083 0.0000 0.0000 -0.0144 -0.0105
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0059 -0.0220 0.0000
15. (1.98305) BD ( 1) C 6 - H 12
( 62.04%) 0.7876* C 6 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.8102
-0.0141 0.2175 -0.0038 0.0003 0.0000
0.0083 0.0000 0.0000 0.0144 -0.0105
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0220 -0.0059 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98096 -0.68186 110(g),107(g),114(v),120(v)
73(v),43(v),109(g),112(g)
42(v),72(v)
2. BD ( 1) C 1 - C 6 1.98097 -0.68200 118(g),106(g),119(v),112(v)
33(v),63(v),109(g),120(g)
62(v),32(v)
3. BD ( 2) C 1 - C 6 1.66532 -0.23794 111(v),116(v),35(v),65(v)
4. BD ( 1) C 1 - H 7 1.98305 -0.51236 118(v),110(v),72(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98097 -0.68202 106(g),113(g),109(v),117(v)
23(v),53(v),114(g),112(g)
52(v),22(v)
6. BD ( 2) C 2 - C 3 1.66532 -0.23794 108(v),116(v),55(v),25(v)
7. BD ( 1) C 2 - H 8 1.98305 -0.51234 113(v),107(v),42(v),22(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98096 -0.68184 110(g),115(g),112(v),119(v)
63(v),33(v),114(g),117(g)
32(v),62(v)
9. BD ( 1) C 3 - H 9 1.98305 -0.51236 106(v),115(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - C 5 1.98097 -0.68202 118(g),113(g),114(v),120(v)
43(v),73(v),117(g),119(g)
72(v),42(v)
11. BD ( 2) C 4 - C 5 1.66534 -0.23795 108(v),111(v),45(v),75(v)
12. BD ( 1) C 4 - H 10 1.98305 -0.51236 118(v),110(v),62(v),42(v)
115(g),113(g)
13. BD ( 1) C 5 - C 6 1.98096 -0.68186 115(g),107(g),109(v),117(v)
53(v),23(v),119(g),120(g)
22(v),52(v)
14. BD ( 1) C 5 - H 11 1.98305 -0.51234 113(v),107(v),52(v),72(v)
115(g),118(g)
15. BD ( 1) C 6 - H 12 1.98305 -0.51236 106(v),115(v),22(v),62(v)
107(g),118(g)
16. CR ( 1) C 1 1.99911 -10.04057 73(v),33(v),110(v),118(v)
120(v),112(v)
17. CR ( 1) C 2 1.99911 -10.04056 43(v),23(v),113(v),107(v)
114(v),109(v)
18. CR ( 1) C 3 1.99911 -10.04057 33(v),53(v),106(v),115(v)
112(v),117(v)
19. CR ( 1) C 4 1.99911 -10.04057 63(v),43(v),118(v),110(v)
119(v),114(v)
20. CR ( 1) C 5 1.99911 -10.04056 53(v),73(v),113(v),107(v)
117(v),120(v)
21. CR ( 1) C 6 1.99911 -10.04057 23(v),63(v),106(v),115(v)
109(v),119(v)
The charge both on all the C atoms and on all H atoms are identical since the electrons on the C-C framework are distributed equally, which is consistent with electron delocalisation.
MO analysis for Benzene Molecule
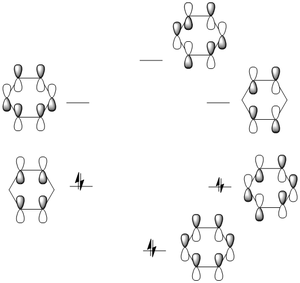
According to Huckel’s rule, fully conjugated, monocyclic systems with (4n+2) pi electrons have a closed shell of electrons all in bonding orbitals and are exceptionally stable are said to be aromatic. Benzene has six pi electrons, thus all its bonding orbitals are occupied. When takes the aromaticity into the consideration of molecular orbitals. The totally bonding orbital is taken into account, in which it has orbitals along both the top face and bottom face of the aromatic system. And this allows the electron delocalisation along the C-C framework, which means each carbon, donates one electron from its molecular orbital into the ring system and there are 6 electrons in the system in total to occupy all the bonding orbitals. Therefore, the concept is in good agreement to the obtained molecular orbital.[6]
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20610
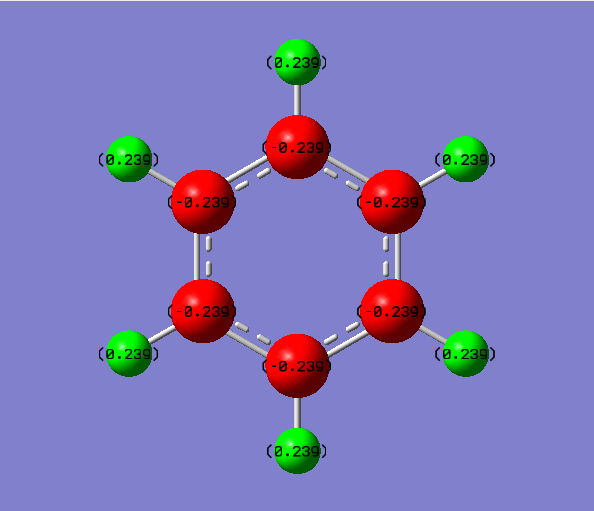
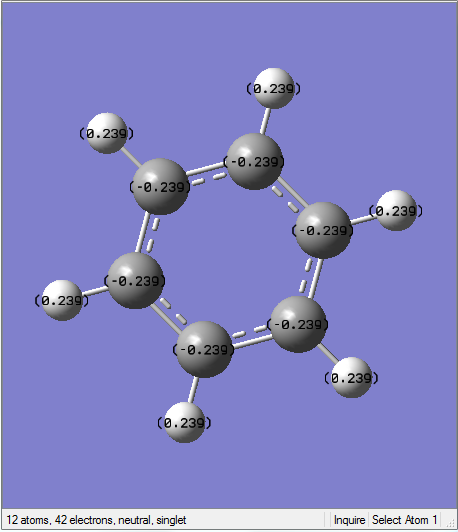
Boratabenzene Molecule
Boratabenzen molecule is isoelectronic to benzene, with one BH- unit replacing the CH unit in benzene molecule. How the characteristics of the ring changes according to replacement of B atom would be discussed as below.
Optimised Boratabenzene Molecule
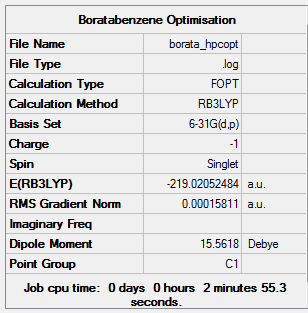
The optimization is completed on the HPC service using B3LYP method with a 6-31G(d,p) basis set.
test molecule |
General Information
| C-C Bond Length | 1.40Å |
| B-C Bond Length | 1.51Å |
B-C bond length is longer than C-C bond length due to the difference in electronegativity and poorer overlap between B atom and C atom. Moreover, the electrons delocalise unevenly and the previous double bond between C-C atom is no longer exist due to the introduction of B atom.
Summary of Optimised Boratabenzene Molecule
Real Output of Optimised Boratabenzene Molecule
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000894 0.001800 YES
RMS Displacement 0.000328 0.001200 YES
Predicted change in Energy=-6.589668D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4053 -DE/DX = -0.0001 !
! R2 R(1,5) 1.3989 -DE/DX = 0.0 !
! R3 R(1,6) 1.0968 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,7) 1.0917 -DE/DX = -0.0001 !
! R6 R(3,4) 1.399 -DE/DX = 0.0 !
! R7 R(3,8) 1.0968 -DE/DX = 0.0001 !
! R8 R(4,9) 1.097 -DE/DX = -0.0001 !
! R9 R(4,12) 1.5137 -DE/DX = 0.0001 !
! R10 R(5,11) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5138 -DE/DX = 0.0001 !
! R12 R(10,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,5) 122.1368 -DE/DX = 0.0001 !
! A2 A(2,1,6) 117.4406 -DE/DX = 0.0 !
! A3 A(5,1,6) 120.4226 -DE/DX = -0.0002 !
! A4 A(1,2,3) 120.4536 -DE/DX = -0.0001 !
! A5 A(1,2,7) 119.7691 -DE/DX = 0.0001 !
! A6 A(3,2,7) 119.7773 -DE/DX = 0.0001 !
! A7 A(2,3,4) 122.1359 -DE/DX = 0.0001 !
! A8 A(2,3,8) 117.4346 -DE/DX = 0.0 !
! A9 A(4,3,8) 120.4295 -DE/DX = -0.0002 !
! A10 A(3,4,9) 115.9452 -DE/DX = 0.0001 !
! A11 A(3,4,12) 120.0798 -DE/DX = -0.0001 !
! A12 A(9,4,12) 123.975 -DE/DX = -0.0001 !
! A13 A(1,5,11) 115.9583 -DE/DX = 0.0001 !
! A14 A(1,5,12) 120.0801 -DE/DX = -0.0001 !
! A15 A(11,5,12) 123.9616 -DE/DX = -0.0001 !
! A16 A(4,12,5) 115.1137 -DE/DX = 0.0 !
! A17 A(4,12,10) 122.4435 -DE/DX = 0.0 !
! A18 A(5,12,10) 122.4428 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0057 -DE/DX = 0.0 !
! D2 D(5,1,2,7) 180.0025 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.004 -DE/DX = 0.0 !
! D4 D(6,1,2,7) 0.0008 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0017 -DE/DX = 0.0 !
! D6 D(2,1,5,12) -0.0009 -DE/DX = 0.0 !
! D7 D(6,1,5,11) 0.0001 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.0009 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0074 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0014 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.0042 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0018 -DE/DX = 0.0 !
! D13 D(2,3,4,9) 180.005 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0041 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0011 -DE/DX = 0.0 !
! D16 D(8,3,4,12) -180.002 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0007 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -179.9999 -DE/DX = 0.0 !
! D19 D(9,4,12,5) -180.0003 -DE/DX = 0.0 !
! D20 D(9,4,12,10) -0.0009 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.0022 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.0017 -DE/DX = 0.0 !
! D23 D(11,5,12,4) 179.9987 -DE/DX = 0.0 !
! D24 D(11,5,12,10) -0.0008 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20620
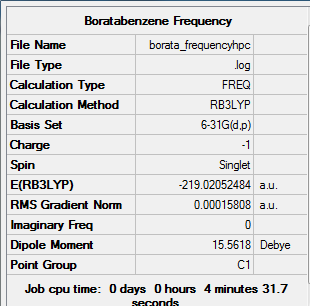
Frequency Analysis for Boratabenzene Molecule
The frequency analysis is carried out using the previous optimisation file and the whole calculation is ran on the HPC service.
Summary of Boratabenzene Frequency
Real Output of Boratabenzene frequency
Low frequencies --- -14.0387 -0.0009 0.0007 0.0008 9.5455 14.6725 Low frequencies --- 371.0132 404.6531 565.1753
The total energy is the same as that of the previous optimisation file.
IR spectrum of Boratabenzene Molecule
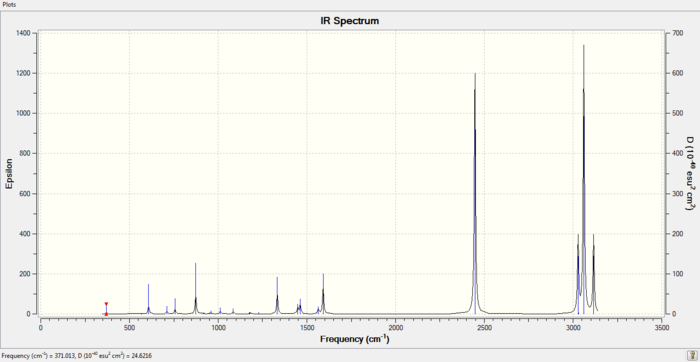
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20623
MO analysis for Boratabenzene Molecule
The MO and NBO analysis is completed using the previous file, involving a full population analysis and full NBOs. The charge range is fixed between -1.000 and 1.000, and how charge distributes is shown below.
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20625
NBO Analysis for Borataenzene Molecule
Charge Distribution of Borataenzene molecule
Specific NBO Charges
Real Output of NBO Analysis for Borataenzene Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.25043 1.99910 4.23721 0.01412 6.25043
C 2 -0.33983 1.99907 4.32693 0.01384 6.33983
C 3 -0.25041 1.99910 4.23719 0.01412 6.25041
C 4 -0.58794 1.99901 4.57715 0.01178 6.58794
C 5 -0.58789 1.99901 4.57709 0.01178 6.58789
H 6 0.17906 0.00000 0.81832 0.00262 0.82094
H 7 0.18563 0.00000 0.81237 0.00200 0.81437
H 8 0.17906 0.00000 0.81831 0.00262 0.82094
H 9 0.18380 0.00000 0.81402 0.00218 0.81620
H 10 -0.09642 0.00000 1.09588 0.00054 1.09642
H 11 0.18380 0.00000 0.81402 0.00218 0.81620
B 12 0.20157 1.99906 2.78777 0.01160 4.79843
=======================================================================
* Total * -1.00000 11.99436 29.91625 0.08939 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.97970) BD ( 1) C 1 - C 2
( 49.96%) 0.7068* C 1 s( 35.51%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6875
-0.0034 -0.4134 -0.0325 -0.0002 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7062
0.0327 0.3753 -0.0141 0.0002 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
2. (1.98270) BD ( 1) C 1 - C 5
( 50.77%) 0.7125* C 1 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0574
0.0311 0.7869 0.0164 0.0006 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 5 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0026
0.0269 -0.8201 -0.0353 -0.0006 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
3. (1.76866) BD ( 2) C 1 - C 5
( 48.13%) 0.6938* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0001
0.0000 -0.0007 0.0000 0.9996 -0.0214
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0001
0.0000 -0.0007 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
4. (1.98570) BD ( 1) C 1 - H 6
( 59.32%) 0.7702* C 1 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7228
-0.0089 -0.4562 0.0100 -0.0004 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 6 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
5. (1.97971) BD ( 1) C 2 - C 3
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.08%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7062
-0.0327 0.3753 -0.0141 0.0003 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 3 s( 35.51%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6875
0.0034 -0.4134 -0.0325 -0.0003 0.0000
-0.0146 0.0000 0.0000 0.0081 -0.0107
6. (1.98507) BD ( 1) C 2 - H 7
( 59.44%) 0.7710* C 2 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 0.0000
0.0000 0.8469 -0.0076 0.0006 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
7. (1.98270) BD ( 1) C 3 - C 4
( 50.77%) 0.7125* C 3 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0574
-0.0311 0.7869 0.0164 0.0006 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 4 s( 32.49%)p 2.08( 67.47%)d 0.00( 0.05%)
0.0000 0.5696 -0.0200 0.0010 0.0026
-0.0269 -0.8202 -0.0353 -0.0006 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
8. (1.76859) BD ( 2) C 3 - C 4
( 48.13%) 0.6937* C 3 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0007 0.0000 0.9996 -0.0214
0.0000 0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0008 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
9. (1.98570) BD ( 1) C 3 - H 8
( 59.32%) 0.7702* C 3 s( 26.88%)p 2.72( 73.07%)d 0.00( 0.05%)
0.0003 -0.5183 -0.0133 0.0012 0.7228
-0.0089 0.4562 -0.0100 0.0003 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
10. (1.98420) BD ( 1) C 4 - H 9
( 59.41%) 0.7708* C 4 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7907
0.0003 0.3470 0.0088 0.0003 0.0000
-0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 9 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
11. (1.96997) BD ( 1) C 4 - B 12
( 66.70%) 0.8167* C 4 s( 42.04%)p 1.38( 57.95%)d 0.00( 0.01%)
0.0000 0.6482 0.0158 0.0012 0.6116
-0.0293 0.4523 0.0090 0.0003 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7056
-0.0393 -0.4070 0.0096 -0.0003 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
12. (1.98420) BD ( 1) C 5 - H 11
( 59.41%) 0.7708* C 5 s( 25.39%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7906
-0.0003 0.3471 0.0088 0.0002 0.0000
0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
13. (1.96996) BD ( 1) C 5 - B 12
( 66.70%) 0.8167* C 5 s( 42.03%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6481 -0.0158 -0.0012 0.6116
-0.0293 -0.4523 -0.0090 -0.0004 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7056
-0.0393 0.4070 -0.0096 0.0003 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
14. (1.98604) BD ( 1) H 10 - B 12
( 55.09%) 0.7422* H 10 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 0.0000
0.0000 0.8172 -0.0016 0.0006 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.97970 -0.46974 108(g),111(g),115(v),118(v)
44(v),63(v),112(g),110(g)
2. BD ( 1) C 1 - C 5 1.98270 -0.46496 119(g),107(g),112(v),110(g)
34(v),120(v),118(g),98(v)
33(v)
3. BD ( 2) C 1 - C 5 1.76866 -0.02907 21(v),22(v),35(v),100(v)
109(g)
4. BD ( 1) C 1 - H 6 1.98570 -0.31413 111(v),119(v),33(v),63(v)
108(g)
5. BD ( 1) C 2 - C 3 1.97971 -0.46976 113(g),107(g),110(v),116(v)
24(v),53(v),112(g),115(g)
6. BD ( 1) C 2 - H 7 1.98507 -0.31743 108(v),113(v),23(v),43(v)
111(g),107(g)
7. BD ( 1) C 3 - C 4 1.98270 -0.46490 117(g),111(g),112(v),115(g)
34(v),120(v),116(g),98(v)
33(v)
8. BD ( 2) C 3 - C 4 1.76859 -0.02905 21(v),22(v),35(v),100(v)
114(g)
9. BD ( 1) C 3 - H 8 1.98570 -0.31413 107(v),117(v),33(v),53(v)
113(g)
10. BD ( 1) C 4 - H 9 1.98420 -0.28848 111(v),117(g),43(v),119(v)
97(v),113(g)
11. BD ( 1) C 4 - B 12 1.96997 -0.31782 113(g),115(v),118(v),116(g)
44(v),43(v),85(v),64(v)
119(g)
12. BD ( 1) C 5 - H 11 1.98420 -0.28849 107(v),119(g),23(v),117(v)
97(v),108(g)
13. BD ( 1) C 5 - B 12 1.96996 -0.31777 108(g),110(v),116(v),118(g)
24(v),23(v),93(v),54(v)
117(g)
14. BD ( 1) H 10 - B 12 1.98604 -0.17251 113(v),108(v),53(v),63(v)
15. CR ( 1) C 1 1.99910 -9.83478 64(v),34(v),119(v),111(v)
112(v),118(v)
16. CR ( 1) C 2 1.99907 -9.82827 44(v),24(v),108(v),113(v)
110(v),115(v),27(v),47(v)
43(v),23(v)
17. CR ( 1) C 3 1.99910 -9.83479 54(v),34(v),117(v),112(v)
107(v),116(v)
18. CR ( 1) C 4 1.99902 -9.79408 44(v),98(v),117(g),111(v)
115(v),97(v)
19. CR ( 1) C 5 1.99902 -9.79408 24(v),98(v),119(g),107(v)
110(v),97(v)
20. CR ( 1) B 12 1.99907 -6.36939 116(v),118(v),113(v),108(v)
53(v),63(v)
21. LP ( 1) C 2 1.14687 0.09689 114(v),109(v),35(g),26(v)
46(v),45(v),25(v)
22. LP*( 1) B 12 0.57264 0.22268 114(v),109(v),100(g),57(v)
Compared to the benzene molecule, the charges on C atoms are no longer equal. The C atoms have a high charge density, whereas the H atoms have a low charge density. When one C atom is being replaced by a B atom, the symmetry of the system is distorted. The H atom attached to the B atom has a negative charge distribution and the carbon atoms attached to B atom have more negative charge distribution. And this is because the electropositive B atoms donate its electron density to its surroundings.
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20625
Pyridinium Molecule
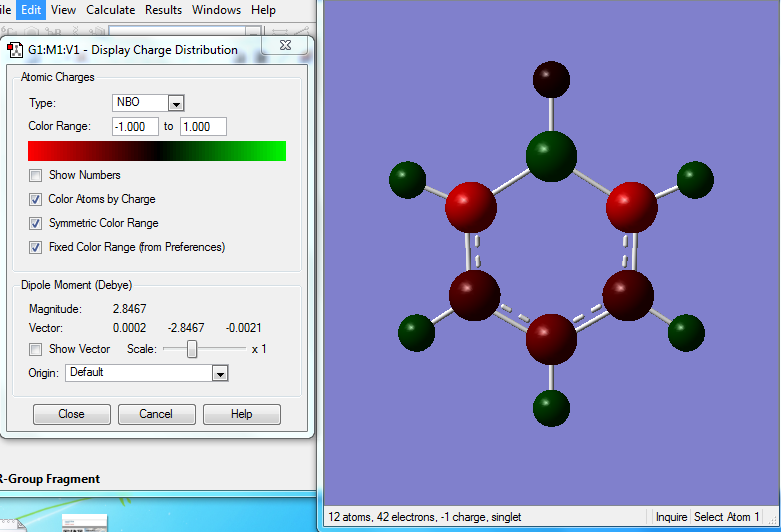
Pyridinium molecule is also isoelectronic to benzene molecule, with one NH+ unit replacing the CH unit on the benzene ring.
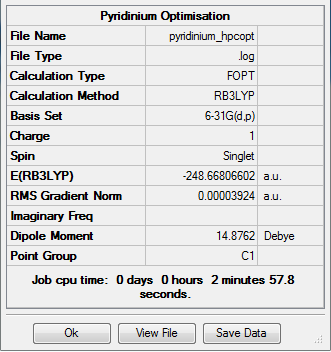
Optimised Pyridinium Molecule
The optimization calculation of pyridinium molecule is completed using a BL3YP method with a 6-31G(d,p) basis set. The symmetry is being switched off.
test molecule |
General Information
| C-C Bond Length | 1.38Å |
| C-N Bond Angle | 1.35Å |
The C-N bond length is shorter than the C-C bond length, but it is shown to be not involved in the conjugated system.
Summary of Optimised Pyridinium Molecule
Real Output of Optimised Pyridinium Molecule
Item Value Threshold Converged?
Maximum Force 0.000065 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000834 0.001800 YES
RMS Displacement 0.000177 0.001200 YES
Predicted change in Energy=-7.095348D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,5) 1.3838 -DE/DX = 0.0 !
! R3 R(1,6) 1.0835 -DE/DX = 0.0 !
! R4 R(2,3) 1.3988 -DE/DX = 0.0 !
! R5 R(2,7) 1.0852 -DE/DX = 0.0 !
! R6 R(3,4) 1.3839 -DE/DX = 0.0 !
! R7 R(3,8) 1.0835 -DE/DX = 0.0 !
! R8 R(4,9) 1.0832 -DE/DX = 0.0 !
! R9 R(4,12) 1.3523 -DE/DX = 0.0001 !
! R10 R(5,11) 1.0832 -DE/DX = 0.0 !
! R11 R(5,12) 1.3524 -DE/DX = 0.0 !
! R12 R(10,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 119.0799 -DE/DX = 0.0 !
! A2 A(2,1,6) 121.4986 -DE/DX = -0.0001 !
! A3 A(5,1,6) 119.4214 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0614 -DE/DX = 0.0 !
! A5 A(1,2,7) 119.9688 -DE/DX = 0.0 !
! A6 A(3,2,7) 119.9697 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.077 -DE/DX = 0.0 !
! A8 A(2,3,8) 121.5031 -DE/DX = -0.0001 !
! A9 A(4,3,8) 119.4199 -DE/DX = 0.0001 !
! A10 A(3,4,9) 123.9283 -DE/DX = 0.0 !
! A11 A(3,4,12) 119.2355 -DE/DX = 0.0 !
! A12 A(9,4,12) 116.8362 -DE/DX = 0.0 !
! A13 A(1,5,11) 123.9339 -DE/DX = 0.0 !
! A14 A(1,5,12) 119.2331 -DE/DX = 0.0 !
! A15 A(11,5,12) 116.833 -DE/DX = 0.0 !
! A16 A(4,12,5) 123.313 -DE/DX = 0.0 !
! A17 A(4,12,10) 118.344 -DE/DX = 0.0 !
! A18 A(5,12,10) 118.343 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0005 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0024 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0016 -DE/DX = 0.0 !
! D4 D(6,1,2,7) -0.0014 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0005 -DE/DX = 0.0 !
! D6 D(2,1,5,12) 0.0021 -DE/DX = 0.0 !
! D7 D(6,1,5,11) -0.0014 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.0011 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0022 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0017 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -179.9993 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0012 -DE/DX = 0.0 !
! D13 D(2,3,4,9) -179.9998 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0013 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0003 -DE/DX = 0.0 !
! D16 D(8,3,4,12) 180.0008 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0014 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -180.0007 -DE/DX = 0.0 !
! D19 D(9,4,12,5) 180.0024 -DE/DX = 0.0 !
! D20 D(9,4,12,10) 0.0003 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.0031 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.001 -DE/DX = 0.0 !
! D23 D(11,5,12,4) -180.0008 -DE/DX = 0.0 !
! D24 D(11,5,12,10) 0.0014 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20698
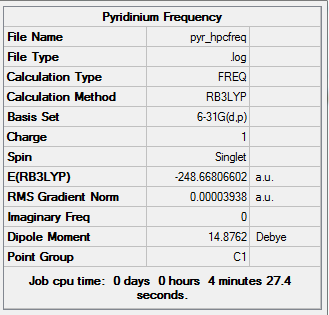
Frequency Analysis for Pyridinium Molecule
The frequency analysis is carried out using the previous optimisation file and the whole calculation is ran on the HPC service.
Summary of Pyridinium Frequency
Real Output of Pyridinium frequency
Low frequencies --- -5.3584 0.0007 0.0007 0.0009 11.0136 14.1165 Low frequencies --- 391.5107 404.4651 620.4190
IR spectrum of Pyridinium Molecule
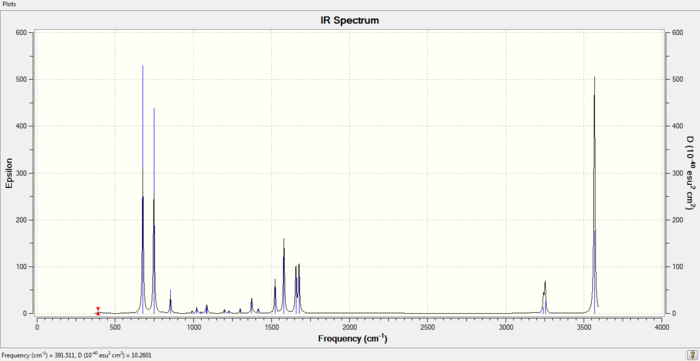
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20728
MO analysis for Pyridinium Molecule
The MO and NBO analysis is completed using the previous file, involving a full population analysis and full NBOs. The charge range is fixed between -1.000 and 1.000, and how charge distributes is shown below.
D-space Link
DOI:10042/to-/https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20729
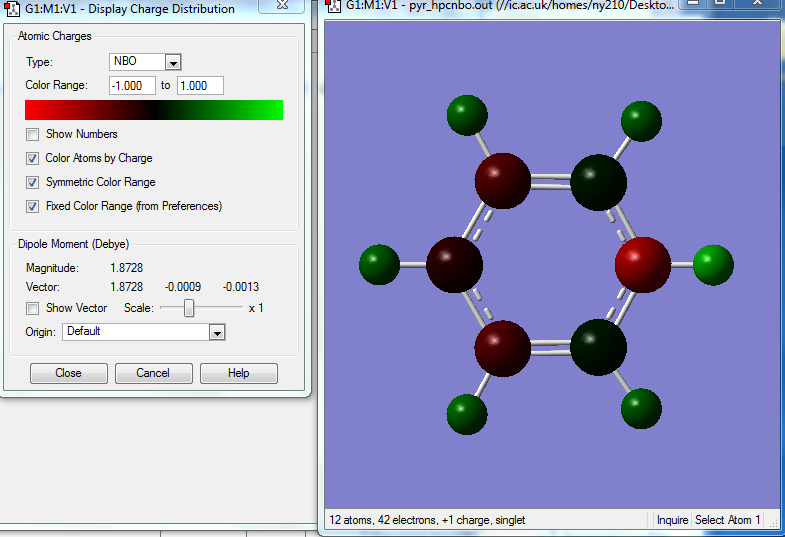
NBO Analysis for Pyridinium Molecule
Charge Distribution of Pyridinium molecule
Specific NBO Charges
Real Output of NBO Analysis for Pyridinium Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.24103 1.99912 4.22859 0.01331 6.24103
C 2 -0.12243 1.99913 4.10943 0.01386 6.12243
C 3 -0.24103 1.99912 4.22859 0.01331 6.24103
C 4 0.07101 1.99918 3.91066 0.01916 5.92899
C 5 0.07099 1.99918 3.91068 0.01916 5.92901
H 6 0.29719 0.00000 0.70178 0.00103 0.70281
H 7 0.29169 0.00000 0.70718 0.00113 0.70831
H 8 0.29719 0.00000 0.70178 0.00103 0.70281
H 9 0.28493 0.00000 0.71397 0.00110 0.71507
H 10 0.48278 0.00000 0.51476 0.00246 0.51722
H 11 0.28493 0.00000 0.71397 0.00110 0.71507
N 12 -0.47622 1.99937 5.46756 0.00929 7.47622
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98249) BD ( 1) C 1 - C 2
( 50.26%) 0.7089* C 1 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4187
-0.0371 -0.6896 0.0068 0.0003 0.0000
0.0122 0.0000 0.0000 -0.0118 -0.0115
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3938
-0.0234 0.7061 0.0290 -0.0003 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
2. (1.98297) BD ( 1) C 1 - C 5
( 49.58%) 0.7042* C 1 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 -0.0011 0.0320 -0.0006 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 5 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 0.0145 0.0331 0.0005 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
3. (1.61445) BD ( 2) C 1 - C 5
( 52.23%) 0.7227* C 1 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9997 -0.0068
0.0000 0.0191 -0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9995 -0.0175
0.0000 -0.0196 -0.0153 0.0000 0.0000
4. (1.97822) BD ( 1) C 1 - H 6
( 64.83%) 0.8052* C 1 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 -0.3985
0.0072 0.7230 -0.0181 0.0003 0.0000
-0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 6 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 0.0116 -0.0208 0.0000
5. (1.98249) BD ( 1) C 2 - C 3
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3931
-0.0235 -0.7065 -0.0289 -0.0003 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 3 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4181
-0.0371 0.6900 -0.0068 0.0003 0.0000
-0.0122 0.0000 0.0000 -0.0119 -0.0115
6. (1.54880) BD ( 2) C 2 - C 3
( 45.73%) 0.6762* C 2 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9997 -0.0036
0.0000 0.0241 -0.0101 0.0000 0.0000
( 54.27%) 0.7367* C 3 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9997 -0.0080
0.0000 0.0086 0.0228 0.0000 0.0000
7. (1.98141) BD ( 1) C 2 - H 7
( 64.64%) 0.8040* C 2 s( 31.07%)p 2.22( 68.90%)d 0.00( 0.03%)
0.0003 -0.5572 -0.0131 0.0007 0.8298
-0.0198 -0.0004 0.0000 -0.0006 0.0000
0.0000 0.0000 0.0000 -0.0153 0.0101
( 35.36%) 0.5947* H 7 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0242 0.0000 0.0000
8. (1.98297) BD ( 1) C 3 - C 4
( 49.58%) 0.7042* C 3 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 0.0004 -0.0320 -0.0006 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 4 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0047 -0.0138 -0.0331 0.0006 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
9. (1.97822) BD ( 1) C 3 - H 8
( 64.83%) 0.8052* C 3 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 0.3991
-0.0072 0.7226 -0.0181 -0.0003 0.0000
-0.0110 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 8 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 -0.0116 -0.0208 0.0000
10. (1.98154) BD ( 1) C 4 - H 9
( 64.26%) 0.8016* C 4 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 -0.4691
0.0193 0.6667 -0.0183 0.0004 0.0000
0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 9 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 0.0128 -0.0209 0.0000
11. (1.98861) BD ( 1) C 4 - N 12
( 36.68%) 0.6057* C 4 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5294 -0.0335 -0.0013 0.4046
0.0563 0.7415 0.0276 -0.0003 0.0000
0.0252 0.0000 0.0000 -0.0184 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 -0.3662
0.0186 -0.7067 -0.0132 0.0002 0.0000
0.0107 0.0000 0.0000 -0.0058 -0.0115
12. (1.82448) BD ( 2) C 4 - N 12
( 28.55%) 0.5343* C 4 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 0.0000 0.0000 0.9991 0.0132
0.0000 0.0102 0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 0.0007
0.0000 -0.0001 0.0000 0.9999 0.0036
0.0000 -0.0128 -0.0077 0.0000 0.0000
13. (1.98154) BD ( 1) C 5 - H 11
( 64.26%) 0.8016* C 5 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 0.4697
-0.0193 0.6663 -0.0183 -0.0003 0.0000
0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 11 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0129 -0.0209 0.0000
14. (1.98861) BD ( 1) C 5 - N 12
( 36.68%) 0.6057* C 5 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4039
0.0563 -0.7418 -0.0277 -0.0003 0.0000
-0.0251 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 -0.3656
0.0187 0.7071 0.0132 0.0003 0.0000
-0.0107 0.0000 0.0000 -0.0059 -0.0115
15. (1.98629) BD ( 1) H 10 - N 12
( 25.41%) 0.5041* H 10 s( 99.88%)p 0.00( 0.12%)
0.9994 -0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.81%)p 2.73( 73.16%)d 0.00( 0.02%)
-0.0002 0.5178 0.0066 -0.0013 0.8553
-0.0091 -0.0004 0.0000 -0.0006 0.0000
0.0000 0.0000 0.0000 0.0115 -0.0106
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98249 -0.90378 118(v),114(v),107(g),63(v)
110(g),109(g),43(v),42(v)
112(g)
2. BD ( 1) C 1 - C 5 1.98297 -0.92650 120(v),112(v),106(g),33(v)
118(g),97(v),109(g),32(v)
119(g),96(v)
3. BD ( 2) C 1 - C 5 1.61445 -0.46666 111(v),117(v),99(v),108(g)
37(v)
4. BD ( 1) C 1 - H 6 1.97822 -0.71855 119(v),110(v),32(v),62(v)
106(g),107(g)
5. BD ( 1) C 2 - C 3 1.98249 -0.90379 115(v),109(v),113(g),54(v)
106(g),114(g),23(v),22(v)
112(g)
6. BD ( 2) C 2 - C 3 1.54880 -0.44892 117(v),108(v),56(v),26(v)
7. BD ( 1) C 2 - H 7 1.98141 -0.71804 107(v),113(v),22(v),42(v)
106(g),110(g)
8. BD ( 1) C 3 - C 4 1.98297 -0.92647 120(v),112(v),110(g),33(v)
115(g),97(v),114(g),32(v)
116(g),96(v)
9. BD ( 1) C 3 - H 8 1.97822 -0.71856 116(v),106(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - H 9 1.98154 -0.75116 119(v),110(v),42(v),113(g)
96(v)
11. BD ( 1) C 4 - N 12 1.98861 -1.06564 119(g),62(v),114(v),43(v)
118(v),63(v),113(g),120(g)
12. BD ( 2) C 4 - N 12 1.82448 -0.56812 108(v),111(v),64(v),46(v)
13. BD ( 1) C 5 - H 11 1.98154 -0.75116 116(v),106(v),22(v),107(g)
96(v)
14. BD ( 1) C 5 - N 12 1.98861 -1.06560 116(g),52(v),109(v),23(v)
115(v),54(v),107(g),120(g)
15. BD ( 1) H 10 - N 12 1.98629 -0.89228 113(v),107(v),52(v),62(v)
16. CR ( 1) C 1 1.99913 -10.26482 33(v),63(v),62(v),118(v)
110(v),72(v),112(v),119(v)
17. CR ( 1) C 2 1.99914 -10.27397 23(v),43(v),107(v),113(v)
114(v),109(v),76(v)
18. CR ( 1) C 3 1.99913 -10.26483 33(v),54(v),52(v),115(v)
106(v),80(v),112(v),116(v)
19. CR ( 1) C 4 1.99918 -10.32333 43(v),119(v),120(v),110(v)
113(g),97(v),84(v),114(v)
20. CR ( 1) C 5 1.99918 -10.32332 23(v),116(v),120(v),106(v)
107(g),97(v),92(v),109(v)
21. CR ( 1) N 12 1.99937 -14.46219 54(v),63(v)
The presence of the electronegative N atom distorts the symmetry of the C-C framework. The H atom and C atoms attached to it have more positve charge distribution due to the electron density withdrawing effect.
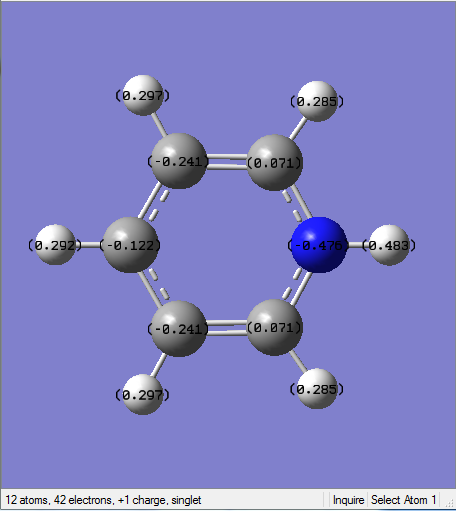
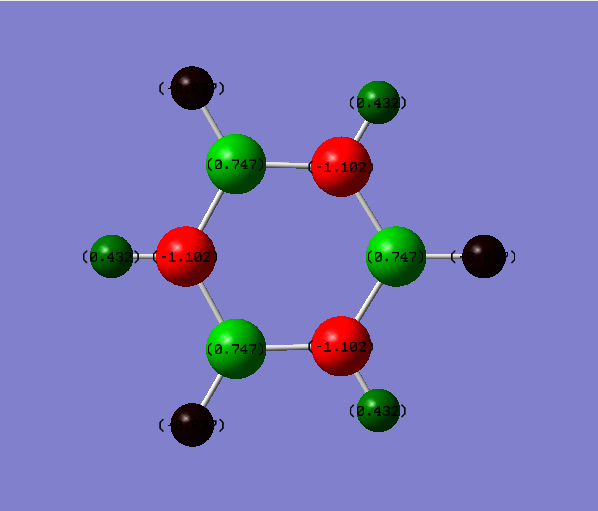
Borazine Molecule
Borazine molecule is isoelectronic to benzene as well, with 3 BH- replacing 3 CH unit and 3 NH+ unit replacing the remaining 3 CH units on the benzene system.
Optimised Borazine Molecule
The optimization is completed on the HPC service using B3LYP method with a 6-31G(d,p) basis set.
test molecule |
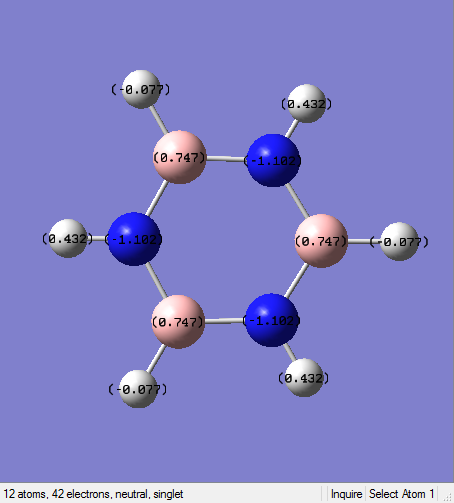
General Information
| B-N Bond Length | 1.43Å |
| N-H Bond Angle | 1.00Å |
| B-H Bond Length | 1.19Å |
The conjugation could not be observed in the Gaussview version. However, this does not mean there is no bond since it is because the bond distance between the atoms may exceed a pre-defined value. All the B-N bond lengths are the same, which gives rise to a conjugated system and electron delocalisation.
Summary of Optimised Borazine Molecule
Real Output of Optimised Borazine Molecule
Item Value Threshold Converged?
Maximum Force 0.000086 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000252 0.001800 YES
RMS Displacement 0.000077 0.001200 YES
Predicted change in Energy=-9.332344D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,9) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,11) 1.0097 -DE/DX = 0.0 !
! R3 R(3,8) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,10) 1.0097 -DE/DX = 0.0 !
! R5 R(5,7) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,12) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4307 -DE/DX = -0.0001 !
! R8 R(7,12) 1.4307 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4307 -DE/DX = -0.0001 !
! R10 R(8,11) 1.4307 -DE/DX = -0.0001 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4306 -DE/DX = -0.0001 !
! A1 A(5,7,10) 121.4399 -DE/DX = 0.0 !
! A2 A(5,7,12) 121.437 -DE/DX = 0.0 !
! A3 A(10,7,12) 117.123 -DE/DX = 0.0 !
! A4 A(3,8,10) 121.4374 -DE/DX = 0.0 !
! A5 A(3,8,11) 121.4415 -DE/DX = 0.0 !
! A6 A(10,8,11) 117.1211 -DE/DX = 0.0 !
! A7 A(1,9,11) 121.4364 -DE/DX = 0.0 !
! A8 A(1,9,12) 121.4407 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1229 -DE/DX = 0.0 !
! A10 A(4,10,7) 118.5641 -DE/DX = 0.0 !
! A11 A(4,10,8) 118.5579 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.878 -DE/DX = 0.0 !
! A13 A(2,11,8) 118.561 -DE/DX = 0.0 !
! A14 A(2,11,9) 118.5609 -DE/DX = 0.0 !
! A15 A(8,11,9) 122.8781 -DE/DX = 0.0 !
! A16 A(6,12,7) 118.5602 -DE/DX = 0.0 !
! A17 A(6,12,9) 118.5628 -DE/DX = 0.0 !
! A18 A(7,12,9) 122.877 -DE/DX = 0.0 !
! D1 D(5,7,10,4) 0.0036 -DE/DX = 0.0 !
! D2 D(5,7,10,8) -179.9997 -DE/DX = 0.0 !
! D3 D(12,7,10,4) -179.9962 -DE/DX = 0.0 !
! D4 D(12,7,10,8) 0.0005 -DE/DX = 0.0 !
! D5 D(5,7,12,6) 0.0003 -DE/DX = 0.0 !
! D6 D(5,7,12,9) 179.997 -DE/DX = 0.0 !
! D7 D(10,7,12,6) -179.9999 -DE/DX = 0.0 !
! D8 D(10,7,12,9) -0.0032 -DE/DX = 0.0 !
! D9 D(3,8,10,4) 0.0007 -DE/DX = 0.0 !
! D10 D(3,8,10,7) -179.996 -DE/DX = 0.0 !
! D11 D(11,8,10,4) -179.9989 -DE/DX = 0.0 !
! D12 D(11,8,10,7) 0.0044 -DE/DX = 0.0 !
! D13 D(3,8,11,2) 0.0035 -DE/DX = 0.0 !
! D14 D(3,8,11,9) 179.9933 -DE/DX = 0.0 !
! D15 D(10,8,11,2) -179.9968 -DE/DX = 0.0 !
! D16 D(10,8,11,9) -0.007 -DE/DX = 0.0 !
! D17 D(1,9,11,2) -0.0057 -DE/DX = 0.0 !
! D18 D(1,9,11,8) -179.9955 -DE/DX = 0.0 !
! D19 D(12,9,11,2) 179.9944 -DE/DX = 0.0 !
! D20 D(12,9,11,8) 0.0045 -DE/DX = 0.0 !
! D21 D(1,9,12,6) -0.0025 -DE/DX = 0.0 !
! D22 D(1,9,12,7) -179.9992 -DE/DX = 0.0 !
! D23 D(11,9,12,6) 179.9975 -DE/DX = 0.0 !
! D24 D(11,9,12,7) 0.0008 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
D-space Link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20788
Frequency Analysis for Borazine Molecule
The frequency analysis is completed using the previous optimisation file and the whole calculation is ran on the HPC service.
Summary of Borazine Frequency
Real Output of Borazine frequency
Low frequencies --- -7.1050 -0.0007 0.0006 0.0011 7.3503 13.0374 Low frequencies --- 288.5924 290.5616 404.2318
IR spectrum of Borazine Molecule

D-space Link DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20791
MO analysis for Borazine Molecule
The MO and NBO analysis is completed using the previous file, involving a full population analysis and full NBOs. The charge range is fixed between -1.000 and 1.000, and how charge distributes is shown below.
D-space Link
DOI:10042/to-/https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20792
NBO Analysis for Borazine Molecule
Charge Distribution of Borazine molecule
Specific NBO Charges
Real Output of NBO Analysis for Borazine Molecule
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43198 0.00000 0.56573 0.00228 0.56802
H 3 -0.07654 0.00000 1.07584 0.00069 1.07654
H 4 0.43199 0.00000 0.56573 0.00228 0.56801
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43199 0.00000 0.56573 0.00228 0.56801
B 7 0.74699 1.99917 2.23864 0.01520 4.25301
B 8 0.74694 1.99917 2.23868 0.01520 4.25306
B 9 0.74698 1.99917 2.23864 0.01520 4.25302
N 10 -1.10242 1.99943 6.09821 0.00478 8.10242
N 11 -1.10241 1.99943 6.09820 0.00478 8.10241
N 12 -1.10241 1.99943 6.09820 0.00478 8.10241
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06889 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98670) BD ( 1) H 1 - B 9
( 54.03%) 0.7351* H 1 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0098 0.0165 0.0000
( 45.97%) 0.6780* B 9 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.4015
0.0137 -0.6802 0.0232 0.0003 0.0000
0.0207 0.0000 0.0000 -0.0114 -0.0098
2. (1.98495) BD ( 1) H 2 - N 11
( 28.08%) 0.5299* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 0.0145 -0.0257 0.0000
( 71.92%) 0.8481* N 11 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 -0.4317
-0.0064 0.7648 0.0114 -0.0007 0.0000
0.0104 0.0000 0.0000 0.0063 0.0119
3. (1.98670) BD ( 1) H 3 - B 8
( 54.03%) 0.7351* H 3 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 -0.0192 0.0002 0.0000
( 45.97%) 0.6780* B 8 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 0.7898
-0.0269 -0.0076 0.0003 0.0003 0.0000
-0.0005 0.0000 0.0000 0.0236 -0.0098
4. (1.98495) BD ( 1) H 4 - N 10
( 28.08%) 0.5299* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 -0.0010 -0.0150 -0.0254 0.0000
( 71.92%) 0.8481* N 10 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
-0.0002 0.4776 -0.0114 0.0006 0.4464
0.0066 0.7563 0.0112 -0.0003 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0119
5. (1.98670) BD ( 1) H 5 - B 7
( 54.03%) 0.7351* H 5 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0094 -0.0167 0.0000
( 45.97%) 0.6780* B 7 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.3883
0.0132 0.6878 -0.0234 -0.0006 0.0000
-0.0202 0.0000 0.0000 -0.0122 -0.0098
6. (1.98495) BD ( 1) H 6 - N 12
( 28.08%) 0.5299* H 6 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 -0.0295 0.0003 0.0000
( 71.92%) 0.8481* N 12 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 0.8782
0.0131 -0.0085 -0.0001 0.0003 0.0000
0.0002 0.0000 0.0000 -0.0122 0.0119
7. (1.98438) BD ( 1) B 7 - N 10
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.8251
-0.0457 0.0304 -0.0355 -0.0003 0.0000
0.0065 0.0000 0.0000 -0.0447 0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7807
-0.0082 -0.0685 -0.0136 0.0003 0.0000
0.0011 0.0000 0.0000 -0.0071 0.0085
8. (1.82090) BD ( 2) B 7 - N 10
( 11.79%) 0.3433* B 7 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 0.9976 -0.0315
0.0000 0.0606 -0.0102 0.0000 0.0000
( 88.21%) 0.9392* N 10 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0006 0.0000 1.0000 -0.0003
0.0000 -0.0028 -0.0037 0.0000 0.0000
9. (1.98438) BD ( 1) B 7 - N 12
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.4002
-0.0540 -0.7222 -0.0208 0.0004 0.0000
0.0365 0.0000 0.0000 -0.0266 -0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.3447
-0.0159 0.7038 0.0000 -0.0004 0.0000
0.0058 0.0000 0.0000 -0.0043 -0.0085
10. (1.98438) BD ( 1) B 8 - N 10
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.4253
0.0090 0.7077 0.0571 -0.0006 0.0000
-0.0413 0.0000 0.0000 -0.0183 -0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.4372
0.0080 -0.6504 0.0138 0.0006 0.0000
-0.0066 0.0000 0.0000 -0.0029 -0.0085
11. (1.98438) BD ( 1) B 8 - N 11
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.4389
0.0079 -0.6994 -0.0573 0.0003 0.0000
0.0420 0.0000 0.0000 -0.0167 -0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.4496
0.0077 0.6418 -0.0139 -0.0003 0.0000
0.0067 0.0000 0.0000 -0.0026 -0.0085
12. (1.82092) BD ( 2) B 8 - N 11
( 11.79%) 0.3433* B 8 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 0.0006 0.0000 0.9976 -0.0315
0.0000 -0.0391 -0.0473 0.0000 0.0000
( 88.21%) 0.9392* N 11 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 1.0000 -0.0003
0.0000 -0.0018 0.0043 0.0000 0.0000
13. (1.98438) BD ( 1) B 9 - N 11
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.8255
-0.0450 -0.0145 0.0364 -0.0002 0.0000
-0.0048 0.0000 0.0000 -0.0450 0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7818
-0.0079 0.0534 0.0138 0.0002 0.0000
-0.0008 0.0000 0.0000 -0.0072 0.0085
14. (1.98438) BD ( 1) B 9 - N 12
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.3862
0.0536 -0.7297 -0.0218 0.0006 0.0000
0.0355 0.0000 0.0000 0.0280 0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.3310
0.0159 0.7103 -0.0003 -0.0006 0.0000
0.0056 0.0000 0.0000 0.0045 0.0085
15. (1.82091) BD ( 2) B 9 - N 12
( 11.79%) 0.3433* B 9 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0006 0.0000 0.9976 -0.0315
0.0000 -0.0215 0.0575 0.0000 -0.0001
( 88.21%) 0.9392* N 12 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 1.0000 -0.0003
0.0000 0.0046 -0.0005 0.0000 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H6B3N3)
1. BD ( 1) H 1 - B 9 1.98670 -0.40393 114(v),116(v),96(v),86(v)
2. BD ( 1) H 2 - N 11 1.98495 -0.61481 115(v),119(v),116(g),118(g)
66(v),56(v)
3. BD ( 1) H 3 - B 8 1.98670 -0.40393 118(v),112(v),86(v),76(v)
4. BD ( 1) H 4 - N 10 1.98495 -0.61481 116(v),114(v),112(g),115(g)
46(v),56(v)
5. BD ( 1) H 5 - B 7 1.98670 -0.40393 119(v),115(v),76(v),96(v)
6. BD ( 1) H 6 - N 12 1.98495 -0.61482 112(v),118(v),119(g),114(g)
46(v),66(v)
7. BD ( 1) B 7 - N 10 1.98438 -0.68871 115(g),111(v),109(g),108(v)
57(v),116(v)
8. BD ( 2) B 7 - N 10 1.82090 -0.27139 117(v),62(v),58(v),35(v)
113(g)
9. BD ( 1) B 7 - N 12 1.98438 -0.68870 119(g),109(v),111(g),106(v)
67(v),118(v)
10. BD ( 1) B 8 - N 10 1.98438 -0.68869 112(g),107(v),109(g),110(v)
47(v),114(v)
11. BD ( 1) B 8 - N 11 1.98438 -0.68872 118(g),109(v),107(g),106(v)
67(v),119(v)
12. BD ( 2) B 8 - N 11 1.82092 -0.27139 120(v),72(v),68(v),27(v)
117(g)
13. BD ( 1) B 9 - N 11 1.98438 -0.68868 116(g),111(v),107(g),108(v)
57(v),115(v)
14. BD ( 1) B 9 - N 12 1.98438 -0.68873 114(g),107(v),111(g),110(v)
47(v),112(v)
15. BD ( 2) B 9 - N 12 1.82091 -0.27140 113(v),52(v),48(v),43(v)
120(g)
16. CR ( 1) B 7 1.99917 -6.65247 115(v),119(v),109(v),111(v)
17. CR ( 1) B 8 1.99917 -6.65247 118(v),112(v),107(v),109(v)
18. CR ( 1) B 9 1.99917 -6.65247 114(v),116(v),111(v),107(v)
19. CR ( 1) N 10 1.99943 -14.13097 57(v),47(v),112(g),115(g)
20. CR ( 1) N 11 1.99943 -14.13097 57(v),67(v),116(g),118(g)
21. CR ( 1) N 12 1.99943 -14.13097 67(v),47(v),119(g),114(g)
The charge distribution of the borazine molecule is symmetric since the charge distributions on the same atoms are equal. The H atoms attached to the N atoms have a lower electron density since the electronegative N atoms pull the electron density, whereas the H atoms attached to the B atoms have a higher electron density since the electropositive B donate the electron density. Moreover, the electron density around the B atoms are reduced due to the electron withdrawing effect from the N atoms.
Discussion
Totally Bonding pi Orbitals
The totally bonding pi orbital is found to be the 17th molecular orbital for benzene molecule. And six C p orbitals are perpendicular to the plane to the benzene ring, which gives rise to a conjugated system with all the p orbitals overlapping with each other in-phase.
| Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|
 |
 |
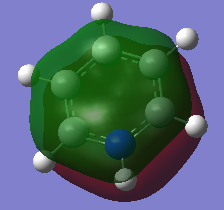 |
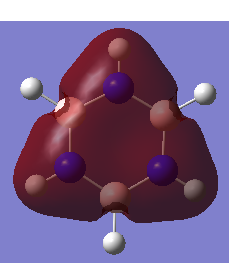
|
From the graphs shown above, the totally bonding pi orbitals are similar for the four isoelectronic molecules since the p orbitals of all the atoms involving in the ring system have good overlap with each other in phase. For benzene, the electron density is delocalised symmetrically on the 6 carbons inside the ring. However, the size of molecular orbitals became different in the region of where heteroatoms are present. This main difference is caused by the difference in electronegativity. For the more electronegative species, the size of p orbitals involved is larger. Thus for pyridinium molecule and borazine molecule, the molecular orbital on the N region is larger. Similarly, the size of p orbitals for the more electropositive species is smaller and the molecular orbital on the B region in boratabenzene molecule and borazine molecule is smaller.
Highest Occupied Molecular Orbital
The HOMO is the 21st molecular orbital for the referencing benzene molecule and has one nodal plane.
| Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|
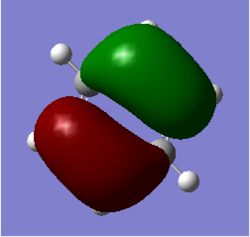 |
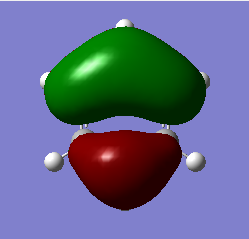 |
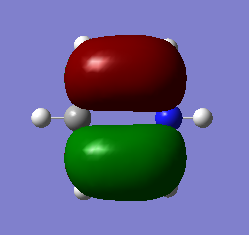 |
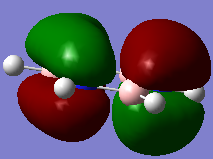
|
The molecular orbitals are the same for all the atoms involved in the C framework for the benzene molecule. All other three isoelectronic compounds show similar patterns of the bonding and antibonding interactions. The N atom on the pyridinium molecule is not involved in the HOMO. Thus the electron-withdrawing effect from electronegative atom is not so obvious and the size of the two lopes is the same. However, the shape of molecular orbital differs from the benzene molecule since only 4 p orbitals are involved in the molecular orbital. The shape of molecular orbital for the borazine molecule differs from the benzene molecule as well since only 4 p orbitals on the alternating B-N framework, two from B atoms and the other two from N atoms are involved. Although the two lopes are the same in size, the molecular orbital does not distribute symmetrically due to the difference in electronegativity between the electronegative N atom and the electropositive B atom. The HOMO of the boratabenzene molecule is quite different from the benzene molecule since the p orbital on the electropositive B atom is involved. Since it is electropositive, it has a higher energy level and could not overlap well with the adjacent C atoms.
Lowest Unoccupied Molecular Orbital
The LUMO is the 22nd molecular orbital for the benzene molecule, which is regarded as a reference. Moreover, the LUMO have two nodal planes and is slightly antibonding.
| Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|
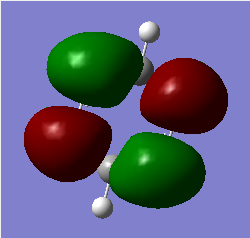 |
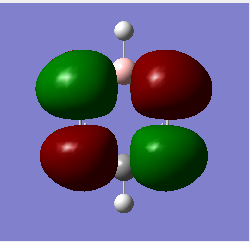 |
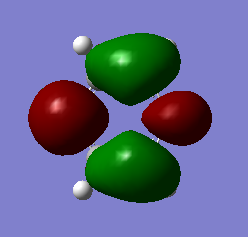 |
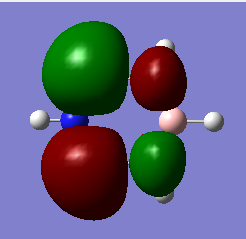
|
The benzene molecule has a LUMO consists of 4 p orbitals on the C atoms. The molecular orbitals are the same for all the atoms involved in the C framework. All other three isoelectronic compounds show similar patterns of the bonding and antibonding interactions.
The boratabenzene molecule has a LUMO consists of 4 p orbitals on the C atoms, without containing the p orbital of the electropositive B atom. Thus the size of molecular orbital looks the same as that of benzene molecule since the electron-donating effect from B atom is small.
The pyridinium molecule has a LUMO consists of 1 p orbital on the N atom and 3 p orbitals on the C atoms. Thus the size of molecular orbital differs a lot from that of benzene molecule. C atoms are more electropositive than the N atom, thus the orbital is larger on the C atoms. The C atom away from the electronegative N experiences less electron-withdrawing effect.
The borazine molecule has a LUMO consists of 4 p orbitals on the alternating B-N framework, two from B atoms and the other two from N atoms. The size of molecular orbital on the electropositive B atom is larger, whereas that on the electronegative N atom is smaller.
In the benzene molecule, all the atomic orbitals are equivalent and would make same contribution to the resulting MO. Since they are all equivalent, the energy matches up and the overlapping is quite good.
However, the equivalence breaks up in the isoelectronic compounds. For the boratabenzene molecule, one C on the benzene ring is replacing by a B atom. B atom is more electropositive than the C atom, and its energy level is higher. The degeneracy is lost, which gives rise to energy mismatch and poorer overlap. Moreover, the stabilisation energy is less. For the pyridinium molecule, one C on the benzene ring is substituted by an N atom. N atom is more electronegative than the C atom, and its energy level lies lower. The degeneracy also loses, which results in energy mismatch, poorer overlap and smaller stabilisation energy. For the borazine molecule, all the C atoms are substituted, in which 3 by B atoms and another 3 by N atoms. The three degenerate B orbitals and three degenerate N orbitals are arranged alternatively, so the degeneracy is lost. The difference in electronegativity is even larger between these two atoms. Thus the overlapping is even poorer and the stabilisation energy is much less.
Referencing
- ↑ J. Blixt, J. Glaser, J. Mink, I. Persson, P. Persson and M. Sandtroem, J. Am. Chem. Soc., 1995, 117 (18), pp 5089-5104
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedbond length - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedLiterature Value of BH3 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedTlBr3 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedformula - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedAromaticity
Cite error: <ref> tag with name "bond length" defined in <references> group "" has no content.
Shriver& Atkins, Inorganic Chemistry, 5th edition, 2010, OUP
Cite error: <ref> tag with name "Literature Value of BH3" defined in <references> group "" has no content.
Kentarou.K et al., J. Chem. Phys., 1987, 87, 2438
Cite error: <ref> tag with name "TlBr3" defined in <references> group "" has no content.
J. E. D. Davies et al., J. Chem. Soc., 1968, 2050
Cite error: <ref> tag with name "formula" defined in <references> group "" has no content.
P. Atkins and J. De paula, Atkins' Physical Chemistry, 9th edition, 2010, OUP
Cite error: <ref> tag with name "Aromaticity" defined in <references> group "" has no content.
Clayden, Greeves, Warren and Wothers, Organic Chemistry, 2001, OUP