User:Zz3116
NH3 molecule
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -56.55776873 a.u.
RMS Gradient Norm = 0.00000485 a.u.
Point Group = C3V
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986258D-10 Optimization completed. -- Stationary point found. ----------------------------
NH3 |
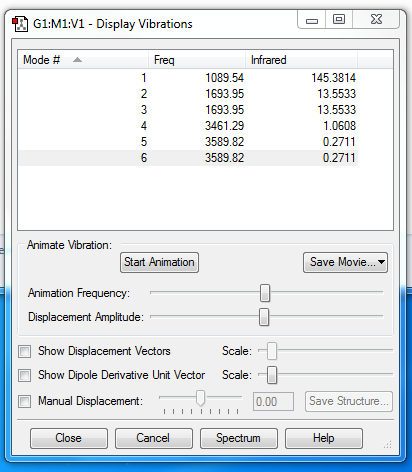
how many modes do you expect from the 3N-6 rule? 6 which modes are degenerate (ie have the same energy)? 2and3,5and6 which modes are "bending" vibrations and which are "bond stretch" vibrations? bending:1,2,3; stretch:4,5,6 which mode is highly symmetric? 4 one mode is known as the "umbrella" mode, which one is this? 1 how many bands would you expect to see in an experimental spectrum of gaseous ammonia? 4
charge on the N atom:-1.125 ;
charge on the H atom:0.375;
expect N:negative
expect H:positive
since N is more electro-negative than H
E(NH3)= -56.55776873 a.u.
2*E(NH3)=-113.1153746 a.u
E(N2)=-109.52412868 a.u
E(H2)=-1.15928020 a.u
3*E(H2)=-3.4778406 a.u
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.11340532 a.u
ΔE=-297.7456677KJ/mol
ammonia product is more stable
N2 molecule
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -109.52412868 a.u.
RMS Gradient Norm = 0.00000060 a.u.
Point Group = D*H
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401004D-13
Optimization completed.
-- Stationary point found.
NH3 |
H2 molecule
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -1.15928020 a.u.
RMS Gradient Norm = 0.09719500a.u.
Point Group = D*H
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
test molecule |
F2 molecule
calculation method=RB3LYP Basis Set=6-31G(d,p) E(RB3LYP)=-199.49825218 a.u RMS Gtadient Norm= 0.09719500 a.u Point Group=D*H
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995024D-08
Optimization completed.
-- Stationary point found.
----------------------------
test molecule |
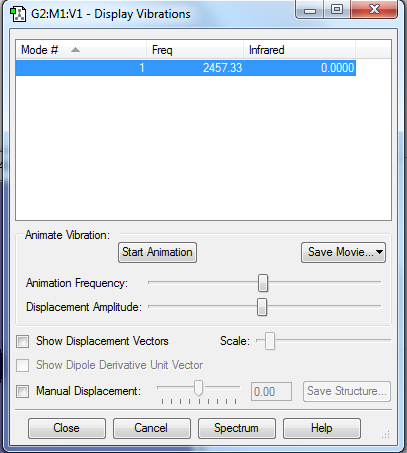
how many modes do you expect from the 3N-5 rule? 1
which modes are "bending" vibrations and which are "bond stretch" vibrations? It is bonding stretch vibration
And it is symmetric
how many bands would you expect to see in an experimental spectrum of gaseous fluorine? 0.Because there is no change in transitional dipole moment.
F charge 0.000, since the two atoms are the same, so no electro-negativity difference.
molecular orbitals of F2





Analysis of F-F bond
The literature value of F-F bond length is 1.42Å.[1] The value we have got from the gaussview is 1.40281Å. They are almost the same. But actually it should be longer than expected due to lone pair repulsion.
- ↑ www.wiredchemist.com/chemistry/data/bond_energies_lengths.html reference.