User:Sw1816
Ammonia Molecule
NH3 optimising
| Question | Answer |
|---|---|
| Molecular name | Ammonia |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -56.55767490 |
| RMS gradient norm/au | 0.00199194 |
| Point group | C3v |
ammonia molecule |
N-H bond length=1.01798 Å H-N-H bond angle=105.741 degrees
The literature value of the N-H bond is 1.01 Å, which matches the optimised ammonia molecule. However, the small uncertainty in the data shows that the structure optimised is not prefect. [1]
Charge distribution N -1.125 H 0.375 (From the optimisation) Charge on the N is expected to be negative and the charge on H is expected to be positive, for the N is much more electronegative than H atom.
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986284D-10
Optimization completed.
-- Stationary point found.
In the 'Item' table, the force value is very small and close to 0, which indicates the gradient of the energy potential to displacement graph is nearly zero. Therefore, the potential energy of the optimised ammonia structure is formed in the most stable structure at the lowest energy.
NH3 molecule vibration
Questions
| Mode | Frequency | Infrared |
|---|---|---|
| 1 | 1089.54 | 145.3814 |
| 2 | 1693.95 | 13.5533 |
| 3 | 1693.95 | 13.5533 |
| 4 | 3461.29 | 1.0608 |
| 5 | 3589.82 | 0.2711 |
| 6 | 3589.82 | 0.2711 |
1. How many modes are expected?
Ammonia molecule has 4 atoms. Ammonia is expected to have 6 vibration modes according to 3N-6 modes.
2. Which modes are degenerate?
The vibration mode 2 and 3 are degenerate modes. The vibration mode 5 and 6 are degenerate modes.
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
The mode 1, 2 and 3 are bending modes and the mode 4, 5 and 6 are stretching.
4. Which mode is highly symmetric?
The first mode is the most highly symmetric viration.
5. Which one is the "umbrella" mode?
The first vibration mode is the 'umbrella mode'.
6. How many bands are expected to see in an experimental spectrum of gaseous ammonia?
There are 2 peaks in the IR spectrum. The highest peak is at 1089.54 cm-1 due to mode 1. The second peak is at 1693.95 cm-1 due to mode 2 and 3. The peaks at 3461.29 and 3589.82 cm-1 do not form a peak due to a low intensity. The mode 1 ,2 and 3 are stretching modes, so the change in dipole moment changes a lot due to change in distance between nitrogen and hydrogen atoms.
Nitrogen Molecule
N2 molecule optimising
| Question | Answer |
|---|---|
| Molecular name | Nitrogen |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -109.52412868 |
| RMS gradient norm/au | 0.00000011 |
| Point group | D∞h |
Nitrogen molecule |
NΞN bond length = 1.10550 Å
The literature value of the NΞN bond is 1.10 Å, which matches the optimised ammonia molecule with small uncertainty.[1]
Charge distribution N 0.000 N 0.0000
Charge distribution of each atom in the molecule is 0.000, because there is no electronegativity difference between these two atoms.
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-1.029996D-14
Optimization completed.
-- Stationary point found.
In the 'Item' table, the force value is 0, which indicates the gradient of the energy potential to displacement graph is zero. Therefore, the potential energy of the optimised nitrogen structure is formed in the most stable structure at the lowest energy.
N2 molecule vibration
Nitrogen molecule have no peak in IR spectrum. The nitrogen only has one frequency due to stretching of the N-N triple bond along the direction of the bond. Therefore, whatever the displacement are changed by the stretching, the changing diploe moment in the vibration of N2 molecule is zero. Thus, the molecule would not absorb energy to vibrate.
Hydrogen Molecule
H2 optimising
| Question | Answer |
|---|---|
| Molecular name | Hydrogen |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -1.17853936 |
| RMS gradient norm/au | 0.00002276 |
| Point Group | D∞h |
Hydrogen molecule |
H-H bond length= 0.74274 Å
The literature value of the H-H bond is 0.74 Å, which matches the optimised hydrogen molecule prefectly. Therefore, the hydrogen atom optimised is very close to the real structure of the hydrogen atom. [1]
Charge distribution H 0.000 H 0.000
Charge distribution of each H atom in the molecule is expected to be 0.000, because these two H atoms have no electronegativity difference between each other.
Item Value Threshold Converged?
Maximum Force 0.000039 0.000450 YES
RMS Force 0.000039 0.000300 YES
Maximum Displacement 0.000052 0.001800 YES
RMS Displacement 0.000073 0.001200 YES
Predicted change in Energy=-2.043043D-09
Optimization completed.
-- Stationary point found.
In the 'Item' table, the force value is 0, which indicates the gradient of the energy potential to displacement graph is zero. Therefore, the potential energy of the optimised hydrogen molecular structure is formed in the most stable structure at the lowest energy.
H2 molecule vibration
H2 molecule has a higher frequency than the N2 molecule. The vibrational frequency is related to force constant and reduced mass. The H2 molecule is lighter than N2 molecule, so the H2 molecule has a higher frequency than nitrogen molecule.
Haber Process
Reaction energy
E(Ammonia)=-56.55767490 Hartrees = -148492.1867615 kJ/mol 2*E(Ammonia)=-113.1153498 Hartrees = -296984.373523 kJ/mol E(Nitrogen)=-109.52412868 Hartrees = -287555.62175417 kJ/mol E(Hydrogen)=-1.17853936 Hartrees = -3094.25532539 kJ/mol 3*E(Hydrogen)=-3.53561808 Hartrees = -9282.76597616 kJ/mol ΔE=2*E(Ammonia)-[E(Nitrogen)+3*E(Hydrogen)]= -0.05560304 Hartrees = -145.99 kJ/mol (to 2 d.p) ΔE(Ammonia)=0.5*ΔE= -72.50 kJ/mol (to 2 d.p)
The reaction energy is negative, so the energy of the ammonia product is lower than the gaseous reactants. The reaction is an exothermic reaction Therefore, the ammonia product is more stable than the nitrogen and hydrogen molecules.
The literature value of the Haber process is -45.8 kJ/mol.[2] The energy of reaction calculated above does not match , but similar to the literature value. This may caused by the optimised structure. The optimised structure is not the real structure and has some uncertainty. Therefore, the calculation does not match perfectly.
Chlorine Trifluoride Molecule
ClF3 optimising
| Question | Answer |
|---|---|
| Molecular name | Chlorine trifluoride |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| E(RB3LYP)/au | -759.46531673 |
| RMS Gradient Norm | 0.00006708 |
| Point group | C2v |
Chlorine trifluoride molecule |
Chlorine trifluoride is a T-shaped molecule. Two F atoms are at axial position forming longer bonds with Cl atom. One F atom is at equatorial position with a shorter bond to Cl atom.
Cl-F bond length 1.65113 Å (one shorter bone), 1.72888 Å (two longer bonds) F-Cl-F bond angle 87.119 degrees(two smaller angles), 174.238(one larger angle)
Charge distribution Cl +1.225 F -0.454(two F atoms), -0.316(one F atom)
The charge distribution og the chlorine is expected to be positive, because the electronegativity of the F atom is higher than Cl atom. In the molecule, each F atom have different distances to the Cl atom. Thus, the F atom with a shorter distance to Cl atom shows less negative.
Item Value Threshold Converged?
Maximum Force 0.000139 0.000450 YES
RMS Force 0.000091 0.000300 YES
Maximum Displacement 0.000845 0.001800 YES
RMS Displacement 0.000552 0.001200 YES
Predicted change in Energy=-1.619042D-07
Optimization completed.
-- Stationary point found.
In the 'Item' table, the force value is close to 0, which indicates the gradient of the energy potential to displacement graph is nearly negligible. Therefore, the potential energy of the optimised ClF3 structure is formed in the most stable structure at the lowest energy.
ClF3 molecule vibration
The frequencies of the ClF3 molecule are low, because the Cl and F atoms are very heavy.
The peak with highest intensity is at 751.75 cm-1. The peak is due to vibration of atoms in a long distance. When the Cl atom vibrates much, the changing dipole moment in the vibration is large. Therefore the mode 6 shows the highest intensity.
ClF3 molecular orbital
The orbital 9 with energy of -1.28172 au (-3365.15612 kJ/mol) shows 3 σ bond. Each of the σ bonds is formed by the overlap of the 3s orbital in Cl atom and the 2s orbital orbitals in F atom. The overlap of the 2s-3s orbitals in phase forms the bonding.
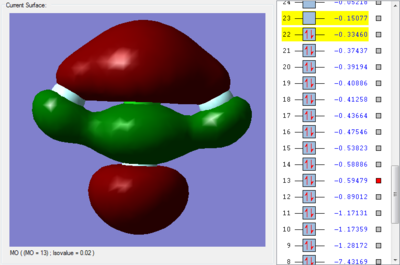
The orbital 13 with energy of -0.59479 au (-1561.62126 kJ/mol) shows 2 π bond and 1 σ bond. The F atom in axial position form π bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals in phase forms the π bonding. The F atom in equatorial position form σ bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals in phase forms the σ bonding.
The orbital 15 with energy of -0.53823 au (-1413.12297 kJ/mol) shows 3 π bond. Each of the F atom form π bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals in phase forms the π bonding.
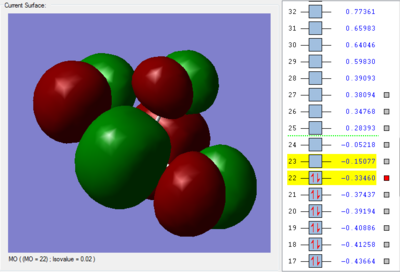
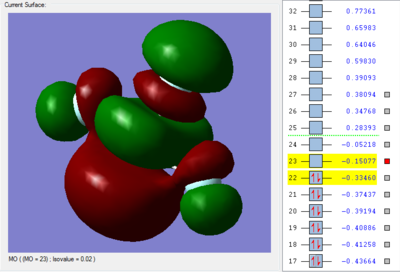
The orbital 22 with energy of -0.33460 au (-878.492367 kJ/mol) shows 3 π* bond. Each of the F atom form π* bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals out of phase forms the π anti-bonding.
The orbital 23 with energy of -0.15077 au (-395.846665 kJ/mol) shows 2 π* bond and 1 σ* bond. The F atom in axial position form π* bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals out of phase forms the π anti-bonding. The F atom in equatorial position form σ* bond with the Cl atom by overlaping of the 2p orbital in F atom and the 3p orbital in Cl atom. The overlap of the 2p-3p orbitals out of phase forms the σ anti-bonding.
Extra calculation
Data used
| Question | Answer |
|---|---|
| Molecular name | Hydrogen |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -1.17853936 |
| RMS gradient norm/au | 0.00002276 |
| Point Group | D∞h |
| Question | Answer |
|---|---|
| Molecular name | Chlorine |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -920.34987886 |
| RMS gradient norm/au | 0.00001378 |
| Point Group | D∞h |
| Question | Answer |
|---|---|
| Molecular name | Hydrogen chloride |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP)/au | -460.80077864 |
| RMS gradient norm/au | 0.00015892 |
| Point Group | C∞V |
H-Cl bond length= 1.28658 Å
Cl-Cl bond length= 2.04168 Å
H-H bond length= 0.74274 Å
Calculation and analysis
The calculation below aims to gain the reaction energy of the reaction between chlorine and hydrogen. the chlorine and hydrogen could react with UV light to undergo a free-radical reaction.
E(Chlorine)=-920.34987886 Hartrees E(Hydrogen)=-1.17853936 Hartrees E(HCl)=-460.80077864 Hartrees ΔEr=2*E(HCl)-[E(Chlorine)+E(Hydrogen)]= -0.07313898 Hartrees ΔE=0.5*ΔEr= -0.03656949 Hartrees = -96.01 kJ/mol (to 2 d.p)
The reaction is an exothermic reaction as the energy of the HCl is Lower than the gaseous reactants. Therefore, the product is more stable. The literature value of the reaction is -93.2 kJ/mol.[3] The optimised data is pretty close to the literature value.