User:Mg5715
NH3, N2, H2 and ClF were looked at in this wiki page and Gaussian was used to optimise and analyse those four molecules. GaussView 5.0 will give summary of the molecule including the energy of molecule. Based on the molecular energy,the reaction energy of the Haber process was calculated. Comparing with the literature value, we know that the value calculated by computer is different from the literature value.
NH3 molecule
The optimisation file of NH3 is linked here
Information
| Parameter | data |
|---|---|
| Molecule name | NH3 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) | -56.55776810 a.u. |
| RMS Gradient Norm | 0.00025207 a.u. |
| point group | C3V |
Item Value Threshold Converged? Maximum Force 0.000419 0.000450 YES RMS Force 0.000274 0.000300 YES Maximum Displacement 0.000894 0.001800 YES RMS Displacement 0.000551 0.001200 YES
NH3 molecule |
Vibration
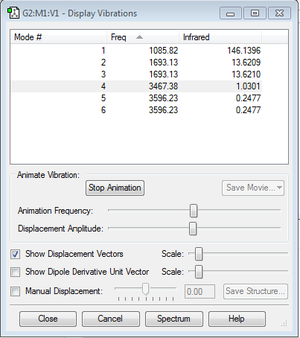
Qustions
1.how many modes do you expect from the 3N-6 rule?
3*4-6=6
2.which modes are degenerate (ie have the same energy)?
mode 2 with mode 3& mode 5 with mode 6
3.which modes are "bending" vibrations and which are "bond stretch" vibrations?
bending: mode 1, mode 2, and mode 3
stretching: mode 4, mode 5, and mode 6
4.which mode is highly symmetric?
mode 4
5.one mode is known as the "umbrella" mode, which one is this?
mode 1
6.how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2. Only if the dipole moment changes while vibrating, it will give a band in spectrum. Mode 4,5,and 6 give small changes in dipole moment, so the intensities are small and will not give bands in the spectrum.
Charge Analysis
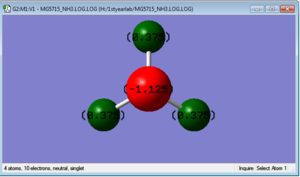
charge on N=-1.125
charge on H=+0.375
Because the overall charge on NH3 should be zero and the electronegativity of N is greater, N will have negative charge and H will have positive charge.
N2 molecule
The optimisation file of N2 is linked here
Information
| Parameter | data |
|---|---|
| Molecule name | N2 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient Norm | 0.00000060 a.u. |
| point group | D∞H |
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
N2 molecule |
Vibration
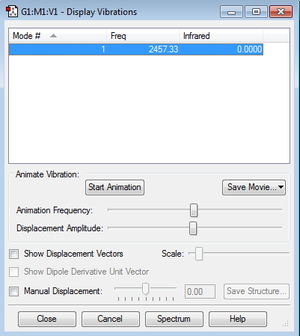
There is only one vibration mode in N2 and it will not give any band in spectrum because the dipole moment does not change.
Charge Analysis
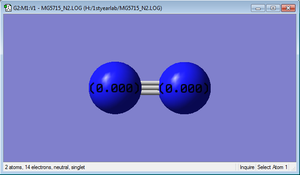
charge on N=0
That is because there in no electronegativity differences between two N atoms. Hence electrons are shared equally between the two N atoms.
H2 molecule
The optimisation file of H2 is linked here
Information
| Parameter | data |
|---|---|
| Molecule name | H2 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) | -1.17853936 a.u. |
| RMS Gradient Norm | 0.00000017 a.u. |
| point group | D∞H |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
H2 molecule |
Vibration
There is only one vibration mode in H2 and it will not give any band in spectrum because the dipole moment does not change.
Charge Analysis
charge on H=0
That is because there in no electronegativity differences between two H atoms. Hence electrons are shared equally between the two H atoms.
Haber-Bosch process
E(NH3)=-56.55776810 a.u.
2*E(NH3)=-113.1155362 a.u.
E(N2)=-109.52412868 a.u.
E(H2)=-1.17853936 a.u.
3*E(H2)=-3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05578944 a.u.=-146.8143158 kJ/mol
Haber-Bosch process is exothermic and this indicates energy level of NH3 is lower than the engery level of N2 and H2.
Literature value=-92.4 kJ/mol[1]
The value calculated by the computer is not same as the literature value which shows that the computer is biased.
ClF molecule
The optimisation file of ClF is linked here
Information
| Parameter | data |
|---|---|
| Molecule name | ClF |
| Calculation Type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) | -559.94269584 a.u. |
| RMS Gradient Norm | 0.00009730 a.u. |
| Dipole Moment | 0.9773 Debye |
| point group | C∞V |
Item Value Threshold Converged? Maximum Force 0.000169 0.000450 YES RMS Force 0.000169 0.000300 YES Maximum Displacement 0.000295 0.001800 YES RMS Displacement 0.000418 0.001200 YES
ClF molecule |
Vibration

Only one brand is showed in the spectrum because it is diatomic molecule and it will give one vibration mode. When the molecule is vibrating, the dipole moment of this molecule is changed because of difference in electronegativities.
Charge Analysis
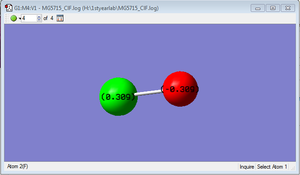
charge on Cl= +0.309
Charge on F= -0.309
Because the electronegativity of F is greater than that of Cl, the charge on F is negative.
MO Analysis
1. 2p-3p πg antibonding orbital

component: 2py of F and 3py of Cl
Energy: -0.32847 a.u.
This molecular orbital is the HOMO (highest occupied molecular orbital)
The 3p orbital of Cl has higher energy level and it is much closer to the energy level of anti-bonding orbital so Cl will have more contribution to the anti- bonding orbital. The bonding π orbital is showed in 3.
2. 2p-3p σu anti-bonding orbital
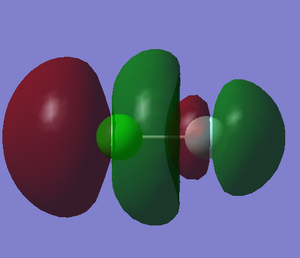
component: 2px of F and 3px of Cl
Energy: -0.12121 a.u.
This molecular orbital is the LUMO (lowest unoccupied molecular orbital)
The 3p orbital of Cl has higher energy level and it is much closer to the energy level of anti-bonding orbital so Cl will have more contribution to the anti- bonding orbital. The bonding σ orbital is showed in 4.
When electron is added in the LUMO, the bond of F and Cl becomes weaker.
3. 2p-3p πu bonding orbital

component: 2py of F and 3py of Cl
Energy: -0.46728 a.u.
The 2p orbital of F has lower energy level and it is much closer to the energy level of bonding orbital so F has more contribution to the bonding orbital. The anti-bonding orbital is showed in 1.
4. 2p-3p σg bonding orbital
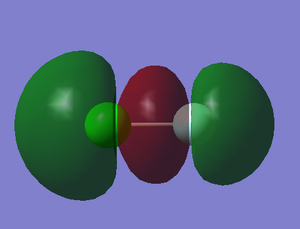
component: 2px of F and 3px of Cl
Energy: -0.52326 a.u.
The 2p orbital of F has lower energy level and it is much closer to the energy level of bonding orbital so F has more contribution to the bonding orbital. The anti-bonding orbital is showed in 2.
5. 2s-3s σg bonding orbital

component: 2s of F and 3s of Cl
Energy: -1.21890 a.u.
The 2s orbital of F has lower energy level and it is much closer to the energy level of bonding orbital so F has more contribution to the bonding orbital.
