TrendsCatalyticActivity
This was an optional experiment for the yr3 comp lab. Now it is discontinued.
Understanding trends in catalytic activity for hydrogen evolution
This is the welcome page for the computational experiment about trends in catalytic activity for hydrogen evolution, part of the third year computational chemistry lab.
This computational experiment has been developed by Dr. Clotilde Cucinotta.
The computational experiment will be from 10:00 to 17:00 on Monday, Tuesday, Thursday and Friday. During these 4 days demonstrators will be available to answer all your questions.
There is an introductory lecture to the lab at 10:00 on the start sate of your session and other lectures in the following days. See the section 'Structure of the Experiment' below.
The deadline is at 12:00 noon on Wed of the week following the experiment.
Work submitted late will be penalised according to the Late Submission Policy.
There is a link set up on Blackboard for submitting the report in PDF format and a .zip folder with all the input files and the output files produced during the experiment, in 3rd Year Chemistry Laboratories (2024 - 2025) / Y3C Third Year Computational Laboratory/;
Please note that the submission boxes will appear only once you have completed the curriculum review.
Your report will be marked out of 20.
The provided template for the report contains a number questions and sub-questions. The final report is expected to contain answers to all these questions.
Questions related to this computational experiment can be directed to the demonstrators, Teams
Their availability is specified below:
| Monday | Tuesday | Thursday | Friday | |
|---|---|---|---|---|
| 10:00-11:00 | Rashid - Kipras | Kipras | Kipras | Kipras |
| 11:00-12:00 | Rashid - Kipras | Kipras | Kipras | Kipras |
| 12:00-13:00 | Rashid - Kipras | Kipras | Kipras | Kipras |
| 15:00-16:00 | Fei | Fei | Fei | Fei |
Outside the time in which the demonstrators are available, all your questions should be posted in the forum on Blackboard.
Introduction
In the electrochemistry course you learned about an important quantity, the exchange current, and how it reflects how viable is the reaction kinetics of an electrochemical transformation.
In this experiment you will predict what is the most efficient metal catalyst for hydrogen evolution by plotting the adsorption energy for hydrogen on different metals versus the measured exchange currents for this reaction. According to the Sabatier principle (named after the chemist Paul Sabatier), high catalytic activity is associated to an interaction between reactants and catalysts, which needs to be neither too strong nor too weak. As a consequence, the plot will have a maximum corresponding to the to the most active catalyst and the ideal adsorption energy: you will observe a so-named volcano plot.
You will learn how to:
- Evaluate the chemisorption energy for an atomic species on a metal;
- generate a volcano plot describing trends in catalytic activity for Hydrogen evolution;
- use the most basic feature of Unix operating system and work remotely on our hpc cluster login.hpc.ic.ac.uk;
- use a quantum-mechanical programme, CP2K, which implements Density Functional Theory (DFT), to calculate the adsorption energies for Hydrogen on the closely packed surfaces of different metals;
- control and evaluate
- the numerical error in your DFT calculations
- the error associated to the selection of simulation parameters such as set of k points used to sample the Brillouin zone and the basis set used to expand the orbitals. Predict the equilibrium lattice parameters for a metal;
- extract from these calculations optimised geometries and energies, and evaluate adsorption energies;
- use a visualization programme to represent relaxation trajectories and optimised configurations.
Please go through the following introductory technical notes, the theoretical notes, and references therein.
Remote Connection
To run this experiment, you will need access to the Imperial College supercomputers. To this end, you need to have Unified Access installed.
It will be useful to set up either Unified Access on your device before the start of the experiment on Monday
- To set up Unified Access on your device you will need to follow this link: using Unified Access
Software
- To generate pictures representing the configurations generated during this experiment you will need to use VMD;
you can download and install Version 1.9.3 (2016-11-30) of VMD following this link
you will need to register
Platforms:
Windows OpenGL (Microsoft Windows XP/Vista/7/8/10 (32-bit) using OpenGL)
MacOS X OpenGL (32-bit Intel x86)
- to plot the graphs produced during this computational experiment you might want to use Anaconda and use Jupiter notebook. Please install them if you will use these tools to make a picture of the Volcano plot.
Instructions
To connect to our cluster your will need to know your college user name and password. You will need to open a terminal (power shell terminal for windows users) and type one of the following lines below:
ssh [your username]@login-ai.cx3.hpc.ic.ac.uk
ssh [your username]@login-b.cx3.hpc.ic.ac.uk
ssh [your username]@login-bi.cx3.hpc.ic.ac.uk
Once you insert your Imperial's password you are on our hpc cluster and can launch your calculations. Since there is no graphical interface available for CP2K code, you will need to submit your calculations from command line using the Unix operating system of our cluster, so the first step is to learn the basics of Unix.
Unix induction tutorial
To carry out the Unix induction tutorial you can use the remote terminal you have launched as instructed above.
To learn all you need about Unix operating system and its most basic commands you can work through the introduction and the first two tutorials of the online course at the University of Surrey.
If you need a bit more practice and want to go more in deep in your knowledge of Unix environment, you can go through the following Unix induction tutorial, developed by Dr. Giuseppe Mallia.
Calculations
When you feel confident with using basic Unix commands (e.g. pwd, cd, ls, cp, mv), you can proceed (again via remote terminal) with running the calculations for the evaluation of the adsorption energy of Hydrogen on different metal substrates.
To this end you will have to evaluate the energy of the clean metal slab , the chemical potential to bring Hydrogen from its reservoir (a Hydrogen molecule in gas phase) to the surface, , and the energy of the system where the hydrogen atom is adsorbed on the metal slab, .
Having these numbers will make possible to evaluate the adsorption energy for Hydrogen on the metal
is the number of H atoms adsorbed on the surface. In our case
Steps:
1. You can download all the input files to carry out your calculations by doing:
> wget https://github.com/ImperialCollegeLondon/VolcanoPlot/archive/refs/heads/main.zip
If you wish, you can also download them on your computer https://github.com/ImperialCollegeLondon/VolcanoPlot/archive/main.zip as a .zip file, but this is not needed to run the calculations on our cluster.
Please avoid to save the zip file in a folder with a name with one or more space (" ").
You will see that you will have downloaded a file, main.zip, which you will unzip doing
> tar -xvf main.zip
This will generate a folder named VolcanoPlot-main, containing among others, a tar file Volcano-input.tar. You need to go to this directory and untar the Volcano-input.tar file:
> cd VolcanoPlot-main
> tar -xvf Volcano-input.tar
Once untared you will find there are several directories. You will to focus initially on:
- Pt Mo Ag Au: Directories with the input files to evaluate the energy of the metal surfaces and the energy of the metal surface with a H atom adsorbed on it.
- H2: directory with the input files to evaluate the energy of the hydrogen molecule
- files: folder containing the basis set and potential files, needed to run the CP2K calculation. Please keep them in this location
2. Go to project folder named as the metal surface it represents. For example, for platinum you will have to write:
> cd Pt
in that folder, you will find two subfolders, named as 'METAL' and 'METAL-H'. You will be able to list these folders doing:
> ls
The folder 'METAL' and 'METAL-H' contain three files each: cp2k.inp, job.pbs and Struct_in.xyz, needed to run the code for the calculation of energy and relaxed atomic structure of the metallic surface under consideration. In particular, cp2k.inp is the input file with the simulation parameters; Struct_in.xyz contains the Cartesian coordinates for the atoms in your system; job.pbs is the script that you will need to submit to run your calculation (see instruction below). You will be able to list them by doing:
> ls METAL/
or
> ls METAL-H/
3. go to one of the subfolders (you may start from the subfolder named METAL)and submit the calculation by doing
> qsub job.pbs
4. check the status of the job by doing:
> qstat
If the job is correctly submitted you will get some information about the number identifying you job (Job ID), the specific 'Class' of calculation, the name assigned to this job (Job Name), the status of your job (it could be running, or queued (Status) and additional comments related to the starting time of your simulation (Comment). Do not worry if the starting time of your simulation is not very close. This information is not final.
5. Take note of the Job Name and the corresponding running directory (the absolute path for your calculation may be something similar to:
/rds/general/user/[YOUR USERNAME]/home/VolcanoPlot-main/[SYSTEM]/[METAL, METAL-H or MOLECULE]/)
6. Wait until the job is finished. You do not need to stay in this directory. You can now go to the other subfolder (if you started from the subfolder named 'METAL' can now do
cd ../METAL-H
and do the steps 2 to 4 to submit another calculation.
7. Once you have submitted the calculations in 'METAL' and 'METAL-H' for the first metal, you can run the calculations for the other metals and for the H molecule, repeating steps 2 to 6.
8.Check from time to time if your calculations are still running by doing
> qstat
If you see that a calculation is not running anymore (its JOB ID will have disappeared from the qstat list), it might mean that the calculation is complete.
9. Go to the directory where the calculation was running. You will see there several files. The most relevant for you are:
- log.out is the main output of your simulation, with information about the convergence of the SCF cycle and
- Volcano-pos-1.xyz is the trajectory file with all the the geometry steps.
- Volcano-RESTART.wfn contains the wavefunctions for your system.
Now copy the files cp2k.inp, log.out and Volcano-pos-1.xyz for Pt METAL-H system and edit them:
What do you observe editing the cp2k.inp file? Can you tell
- What is the basis set used?
- What kind of calculation did you perform?
- What is the target accuracy for force calculation?
- What is the target accuracy for the SCF convergence?
- Can you extract from the input the lattice parameters for your supercell?
- Is this an orthorombic cell?
- How many atoms do you have in your system?
- What is the hydrogen coverage in your system?
What do you observe editing the log.out file?
- How many self consistent cycles to converge for every geometry optimization step?
- What is the number of electrons and the number of occupied orbitals in your system?
- Did your calculation converge?
To extract the final energy corresponding to the optimised structure for your system you will have to do:
> grep "ENERGY| Total FORCE_EVAL ( QS ) energy (a.u.):" log.out
After that, record the last line of the output. So, for instance, if you are analysing the energy of a metal surface where a H atom has been adsorbed, you will get ; if you are analysing the energy of a clean metal surface, you will get
10. Repeat point 9 for all the systems you have simulated and make a table with the measured exchange currents as per table below and Hydrogen chemisorption energies for all systems.
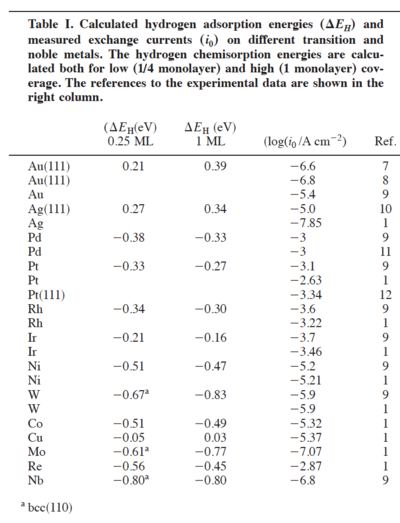
Now make a plot of the exchange currents versus free energy for Hydrogen adsorption.
Do you see a Volcano? If yes, please fit the left and right part of it with two straight lines and create a picture. This is the main picture of your report.
Based on your volcano plot can you tell which metal is the best catalyst for Hydrogen evolution and why?
in this part of the experiment you will:
1. evaluate the error on energy and energy differences associated to the selection of some of the simulation parameters such as kpoints and basis set.
2. evaluate the equilibrium lattice parameter for all the metals you have studied.
You will first evaluate the error on the total energy associated to the Monkhorst-Pack k point set used to describe the Pt surface.
To this end you will have to calculate the total energy of your Pt surface using different k point grids (with a density ranging from 3 3 1 to 15 15 1) and evaluate the differences in total energy with respect to the most accurate set used. Please note that we assume that the (15 15 1) set samples the Brillouin zone accurately enough to be considered our exact reference total energy.
To do this, you will need to untar the file kpoints_unitcell-input.tar you have downloaded with wget. Once done, you will need to run the script kpoints_inputs.sh, by doing:
> ./kpoints_inputs.sh
This will create a separate input directory for every k point set and submit a calculation from that directory. You can check that you have actually submitted all the calculations by using the qstat command.
Once all your calculations have converged, please plot a graph of the energy versus the density of the Monkhorst-Pack grid used; The zero of the energy scale in this graph should be represented by the value of the energy calculated with the most accurate k point set. Please, create a figure with this trend and and include it in your report.
What do you observe?
- You will then study the dependence of Hydrogen chemisorption energies on k-points set and basis set used to describe a 2x2 Pt slab with three layers. This study will be done for two different H coverages, namely 1/4 and 1 ML.
Your input data are contained in the files Pt-basis-2x2-input.tar and pt-kpt-2x2-input.tar. You will need to untar these files and submit the calculations from every generated directory. What is contained in every directory should be straightforward at this stage. Please ask your demonstrator if you have doubts.
Once your calculations have converged, please study the error on H chemisorption energy versus k point set, for both 1/4 and 1 ML coverages, and do the same for H chemisorption energy versus basis set. You will also need to study how the total energies vary with kpoint and basis sets.
- What do you observe? How the error in the evaluation of Hydrogen chemisorption energy versus kpoint and basis sets compares with the error in the evaluation of total energy in this system?
Please create vith VMD a figure representing both H covered surfaces and also create figure for the above trends. These figures will need to be included in your report.
2.
- You will finally evaluate the equilibrium lattice parameters and bulk moduli for Ag, Au, Pt and Mo by fitting the energy versus Volume curve using the Murnagghan equation of state.
For every metal, you will have to calculate the total energy for several Volumes of your cell around the equilibrium volume for your cell. In practice, for every metal you will to run calculations with slightly rescaled coordinates and cell parameters. You will then need to fit these point and find the minimum of the associated Murnagghan curve.
The input data to run the associated calculations are contained in the 'metal-murn-input.tar' file. For each metal ('Ag', 'Au, 'Mo' and 'Pt'), run the calculations using the provided script 'latt_inputs.sh' by typing:
> ./latt_inputs.sh
This will create new directories and input files for different values of lattice parameters; the script will also automatically submit the calculations for each of these lattice parameters. Once all the calculations are finished, use the script 'latt_analyse.sh'. This script will collect the energies vs lattice_parameter data and store them in the 'lattice_data.ssv' file. The script will also create the file 'MURN.inp' which you will need to fit the data using the Murnagan equation of state.
To fit the data you will produce, you will first compile the 'murn-new.f' Fortran file using
> gfortran murn-new.f -o murn-new.x
After this, move the compiled 'murn-new.x' executable to each metal's folder and run it using:
>./murn-new.x < MURN.inp > MURN.out
You will now get the values of equilibrium lattice parameter and bulk modulus by typing:
> grep -A3 "alat=" MURN.out
which should give the following output
alat= 7.48014 b0= 0.8148 b0p= 4.500 E0= -8.379061
7.4801 418.53180 0.81482 4.50041 -228.01101 -16.75850
3.9583
For this example, the equilibrium lattice vector is 3.9583 Angstrom and the bulk modulus is 0.8148 MBar.
For one representative system please plot a graph showing how the total energy varies with the cell volume. How the obtained values compare with experiments?
Please check the materials databases
materials databases aflow library
Finally, untar the file Pt-H-relaxation-input.tar with the input to obtain a relaxation trajectory for Pt slab. Please submit the calculation and visualise the trajectory with VMD. What do you observe?
Storage space
There's a designated storage directory for your use on the cluster. You can find it at
/rds/general/user/[your college username]/projects/complab2021/live/
If you'd like to use this repository, please create your own directory within it. Remember, This is a shared space, so be careful not to delete files that don't belong to you.
Please note that access to the storage space will be removed after submission.
Structure of the experiment
Try to learn to use the Unix environment by Monday at the latest, to have submitted all the calculations by Tuesday and to have finished to run all the calculations by Thursday. It will take time to analyse the results and understand all the new concepts that you will learn in this lab. You should be able to complete your experiment and to write your report by Friday.
Please note that the queueing system of our hpc cluster sometime doesn't allow to predefine the time when a calculation will run. If your calculations won't have finished to run by Thursday morning, the relevant output files to proceed with the analysis will be provided on Thursday morning.
Detailed timeframe
Monday (morning session): 10:00 - 11:00 am: Short introduction to the experiment + Unix induction tutorial. Practise the use of Unix environment and its most basic commands.
Monday (afternoon session): 2:00-2:30 pm short practical introduction to the steps to carry out in the the experiment. Submit your calculations and analyse your files.
Tuesday (morning session): 10:00-11:00 am Lecture: introductory technical notes. Analyse your files and start studying trends.
Tuesday (afternoon session):Analyse trajectories; Study trends.
Thursday (morning session): 10:00-11:00 am: Lecture: theoretical notes . Checkpoint for progress. Visualise your output with VMD.
Thursday (afternoon session). Study trends. Evaluate error. Create the Volcano plot.
Friday (morning session): 10:00-11:00 am: Q&A and short VMD tutorial. Study trends; report write-up;
Friday (afternoon session): report write-up.
Your ability to use HPC resources will end on the Friday of your computational lab week, so make sure you've completed all calculations by then.
Your report is due on Wednesday of the second week following your lab week at 12 noon. This extended deadline allows time for clarifying doubts before submitting.
- Python
- Thermodynamics
- Electrochemistry
Good skills on Unix operating system is not a requirement. All the scripts are written in a way where you only need to submit the script executing the commands. However, feel free to edit and play with the scripts.
New contents
- Unix operating system
- Structural optimizations
- Periodic boundary conditions
- Evaluation of adsorption energies
- Volcano plots and Sabatier principle
Submission of the report
You will need to submit
- The report as a PDF document, via Turnitin in Blackboard.
- all the input and output files you produced in your calculation via Blackboard, as a .zip folder.
Please note that the submission boxes on Blackboard will appear only once you have completed the curriculum review.
Write up
The report structure will consist of three sections:
- Introduction/Summary (Half-page)
- Questions & answers (No page limit)
- Conclusions (Half-page)
Tips to write a report:
- The golden rule: Aim for clarity
- Structured statements that flow in a logical manner.
- Good use of diagrams and appropriate level of theory.
- Careful choice of content.
- Keep your language clear and simple.
- Label all tables and figures. Labels should be self-contained, which means that tables and figures should be interpretable by themself.
- Appropriate referencing of figures and tables.
- Cite previous works (with an accepted citation style) whenever is appropriate.
Introduction/Summary:
- The purpose of the Introduction/Summary is to put the reader in the context of the experiment and to explain how the experiment was carried in the lab. It may contain a brief review of previous research, why the research was undertaken, an explanation of the techniques and why they are used and why it is important in a broader context.
Questions & Answers:
- There are a number of questions in the lab script that need to be answered in this section of the report.
- Depending on the nature of the question, it might be appropriate to use figures or tables to give a proper answer.
- It is highly encouraged to rationalise the answers.
- Please answer the provided questions in the given order. Be sure to mention which question you're answering. Endeavour (e.g. attempting to answer all questions) is worth up to 2 out of 20 points for your evaluation.
Conclusions:
- The Conclusions gives a general description of the results and findings and it should be related back to the Introduction. If appropriate, suggest improvements or additional experiments.
Mark Scheme
The break-down for the marks for this lab are as follows:
| Introduction/Summary | 20% |
| Questions & Answers | 60% |
| Conclusions | 20% |
Plagiarism
Submissions are checked for plagiarism. External images may be used if correctly cited, but it's always better to create your own.
Demonstrators
The demonstrators will be available via Microsoft team channel. Feel free to contact them at the times specified above.
Outside the time in which the demonstrators are available, all your questions should be posted in the forum on blackboard.
Related literature
- W. Schmickler and E. Santos, Interfacial Electrochemistry, 2nd Edition, Springer
- P. W. Atkins Physical Chemistry, Oxford University Press (one edition of your choice)
- Psi-k Scientific Highlight of the Month. Theory of solid/electrolyte interfaces, Axel Gross, Univerisy of Ulm
- R. Martin, Electronic structure : basic theory and practical method - Cambridge, UK ; New York : Cambridge University Press 2008 - ebook available in the library.
- Nørskov, Jens Kehlet; Bligaard, Thomas; Logadottir, Ashildur; Kitchin, J.R.; Chen, J.G.; Pandelov, S.;
Stimming, U., Trends in the exchange current for hydrogen evolution, Journal of The Electrochemical Society, 152 (3) J23-J26 (2005)
- Allen Tildesley, Computer Simulations of Liquids, Oxford University press (2009).
VMD and Periodic Boundary Conditions
On Thursday 2pm there will be a lecture about visualization of atomic structures with of VMD and the role of Perodic Boundary Conditions.
VMD is a visualisation programme which will make possible to visualise among other things, the atomic structure of your surface and the cell used to represent it. and create pictures with them.
To download VMD visualiser follow this link for Version 1.9.3 (2016-11-30) Platforms: Windows OpenGL (Microsoft Windows XP/Vista/7/8/10 (32-bit) using OpenGL)
You will need to register. Registration is free.
Winscp
Winscp can be used to transfer files from and to the hpc platform to and from your computer.
To download winscp you will need to follow this [wincp https://sourceforge.net/projects/winscp/ scp link]
Alternative instructions for Remote Connection
Please check with the demonstrators first.
if you are not able to connect to our hpc cluster using your browser or you could not install on your computer VMD or winscp:
- connect to an Imperial computer following this Imperial remote access link
You will need to follow the instructions and use your username and password.
Once connected you will launch Imperial Apps Anywhere , looking for https://softwarehub.imperial.ac.uk/ in your browser, and look for winSCP and VMD therein.
- Click on the link in the email with subject "Register for Lab - IC_Chemisty_yr3_UK".
You can download files to your computer via command line opening a Linux terminal (or a terminal emulator) on your personal computer.
To this end you can follow this link
