Talk:Modxl9814
JC: General comments: All tasks answered, and results look good. Make sure that you understand the background theory behind these questions and use it to explain your result, and that your explanations don't contradict what your results show.
Theory
Velocity Verlet Algorithm
One way to solve Newton's Second law F=ma is the velocity-Verlet algorithm. By using a Taylor expansion,the atomic positions, velocities and accelerations can be approximated at time t with good precision. The position of atom i, at time t, is denoted by and the velocity of the atom at time t is denoted by . Position at the next timestep can be expressed by:
A single timestep is expressed as:
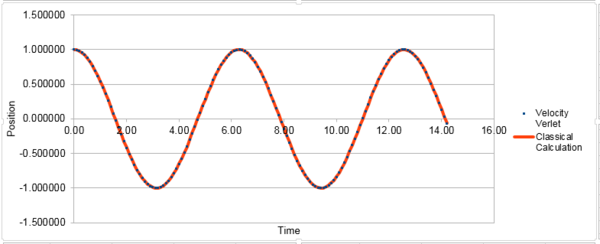
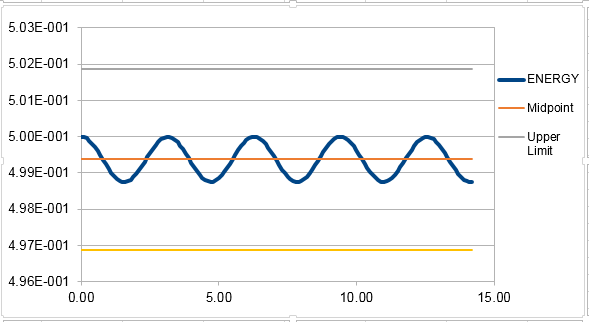
The classical harmonic oscillator can be describe by . The errors oscillate through 5 peaks in the simulated time. The plot of the total energy vs. time of the simulated system:

The cumulative error over a constant interval of time is given by:
Thus, it can be seen from this equation that the relation between the maxima of the error of the Velocity-Verlet algorithm and is quadratically increasing. The graph of the maxima of error vs. time therefore can be fit into the quadratic equation in figure 2.
JC: Good additional research, but a linear fit would have been sufficient here.
The total energy of the oscillating system is the sum of the kinetic energy and the potential energy, with and . In this case, m=1 and k=1, therefore the equation is .
 |
 |
In order for the total energy not to change by more than 1% over the course of the simulation, the timestep needs to be 0.2. It is important to monitor the total energy of the system to ensure that energy conservation is obeyed, the same as the real system.
JC: Good choice of timestep.
Atomic Forces
The Lennard-Jones potential can tell the potential energy of the interaction between two uncharged atoms. It can be expressed in (12,6) form:
In this equation, is the potential well depth, is the distance where the potential between the pair of particles is zero and r is the distance between the pair of particles.
As force is the negative derivative of potential energy, the equation of force in terms of the Lennard-Jones potential is:
When the potential energy is zero, , therefore by substitution we can get:
The equilibrium is reached when the resultant force is zero, therefore:
Divide both sides by :
Therefore, the equilibrium separation is:
The LJpotential at is:
, therefore:
JC: All maths correct.
Periodic Boundary Conditions
. The density of water= under standard consitions (298K, 1atm). So the total mass of 1 mL water= 1g. The number of moles of water molecules=
Therefore the total number of molecules in 1 mL of water=
The volume of 10,000 molecules of water=
The initial position of atom is . After it moves along the vector , the position should be . Applying the periodic boundary of to , the position should be .
Reduced Units
The well depth=
, therefore
JC: All calculations correct.
Equilibration
Creating the simulation box
Giving atoms random starting coordinates may make two atoms generated too close together. This will cause the two atoms to collide and arise the repulsion between the two atoms. The repulsive force between the atoms will drive them apart, leading to increase in the potential energy of the system and making it very unstable.
JC: High repulsive forces can cause the simulation to crash.
A face-centered cubic lattice has 4 lattice points per unit cell. The side length of the cubic unit cell=
If 1000 unit cells were generated by the create_atoms command, 4000 atoms would be generated for a FCC lattice.
Setting the properties of the atoms
Mass 1 1.0
This means the mass of the single type of atom is 1.0.
Pair_style lj/cut 3.0
"Pair_style" indicates that the interaction is pairwise interaction. "lj.cut" describes the standard 12/6 Lennard-Jones potential without a Coulombic pairwise interaction. "3.0" indicates that the global cutoff for atoms is at 3.0.
Pair_coeff * *1.0 1.0
"pair_coeff" specifies the pairwise force field coefficients. The two asterisks indicate that the command will apply to all atoms.
If and are specified,the Velocity-Verlet algorithm will be used for this simulation.
JC: Why do we use a cutoff? What are the forcefield coefficients in your simulations, considering that you are using a Lennard-Jones potential?
Running the simulation
The purpose of defining variable is that we don't need to manually change the numerical timestep each time the timestep needs to be changed. This reduces the human errors that may occur as the timestep only needs to be changed once to the value defined.
JC: Using variables means that all commands in the simulation that depend on a certain value, like the timestep in this case, are updated automatically when that value is changed.
Checking equilibration
 |
 |
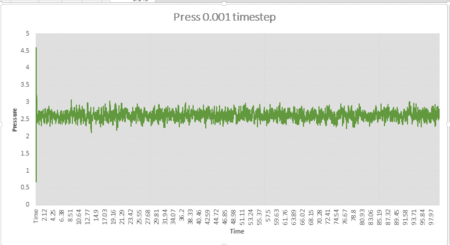 |
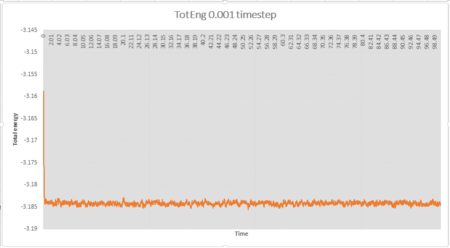
The simulation reaches equilibrium at 0.001 timestep as pressure and temperature become constant. It can be seen from pressure and temperature data that the simulation reaches equilibrium at t=0.29.The average pressure value is about 2.61 and the average temperature value is about 1.26.

It can be seen from Fig 5 that the total energy produced by 0.0025 timestep are very close to those produced by 0.001 timestep. Simulations at 0.0075 and 0.01 also reach equilibrium but the total energies are higher than those produced by 0.001 timestep, thus these timesteps are not very accurate. Therefore the largest timestep to get acceptable results is 0.0025 and the worst choice is 0.015 timestep as the simulation doesn't reach equilibrium.
JC: Good choice of timestep, average energy should not depend on the timestep, making timesteps above 0.0025 poor choices.
Running simulations under specific conditions
Barostat and Thermostat
In the system with N atoms, each with 3 degrees of freedom:
By multiplying every velocity by and substituting T with we can get the second equation:
By substituting (2) we can get:
JC: Correct.
Examining the Input Script
The numbers 100 1000 100000 indicate the timesteps the input values will be used to compute the averages of density, pressure and temperature. For this simulation, the average will be calculated using values produced by timestep 100,200,...100000. Therefore, 1000 values will be used to calculate the average. The following line tells LAMMPS to run the simulation for 100000 timesteps. 0.0025 timestep will be used. Therefore 250 time units are simulated.
JC: Be more specific about what each of the three numbers refers to.
Plotting the Equations of State
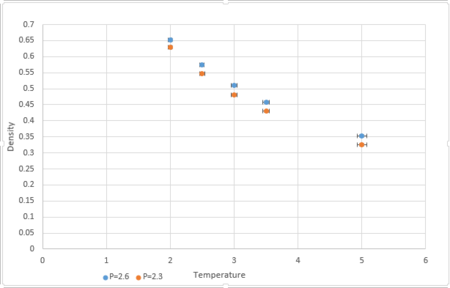 |
 |
Simulations were conducted at temperatures 2,2.5,3,3.5,5 and pressures 2.3 and 2.6.
Density can also be calculated by Ideal Gas Law through the following:
As , and [2], by substitution we can get:
Fig.7 shows that the simulated density is much lower than the density obtained by the Ideal Gas Law. This is because the Ideal Gas Law assumes that the molecules do not interact with each other and the repulsive force between the molecules is zero.This means that the particles in Ideal Gas system can be compressed to great extent, making the volume occupied very small for a given volume. Therefore the density is higher. In the Lennard-Jones model, the molecules will interact with each other and the repulsive force is greater when the distance between the molecules is smaller. Therefore for a given volume the molecules will rather stay far apart and the density is lower .
It can be seen from Fig.7 that the discrepancy increases with pressure. This is because at lower pressure, provided that the volume is large enough, the intermolecular distance is larger and the density will not change a lot by the distance between the particles.
JC: Good, how does the difference between the ideal gas and siulation results change with temperature?
Heat Capacity Calculation
In the NVT ensemble, pressures (0.2,0.8) and temperatures (2,2.2,2.4,2.6,2.8) were used to calculate the heat capacity by using the following equation:
The code to run the simulation in the NVT ensemble is as following:
variable density equal 0.2
### DEFINE SIMULATION BOX GEOMETRY ###
lattice sc ${density}
region box block 0 15 0 15 0 15
create_box 1 box
create_atoms 1 box
### DEFINE PHYSICAL PROPERTIES OF ATOMS ###
mass 1 1.0
pair_style lj/cut/opt 3.0
pair_coeff 1 1 1.0 1.0
neighbor 2.0 bin
### SPECIFY THE REQUIRED THERMODYNAMIC STATE ###
variable T equal 2.0
variable p equal 4
variable timestep equal 0.0025
### ASSIGN ATOMIC VELOCITIES ###
velocity all create ${T} 12345 dist gaussian rot yes mom yes
### SPECIFY ENSEMBLE ###
timestep ${timestep}
fix nve all nve
### THERMODYNAMIC OUTPUT CONTROL ###
thermo_style custom time etotal temp press
thermo 10
### RECORD TRAJECTORY ###
dump traj all custom 1000 output-1 id x y z
### SPECIFY TIMESTEP ###
### RUN SIMULATION TO MELT CRYSTAL ###
run 10000
unfix nve
reset_timestep 0
### BRING SYSTEM TO REQUIRED STATE ###
variable tdamp equal ${timestep}*100
variable pdamp equal ${timestep}*1000
fix nvt all nvt temp ${T} ${T} ${tdamp}
run 10000
reset_timestep 0
### MEASURE SYSTEM STATE ###
thermo_style custom step etotal temp press atoms density vol
variable dens equal density
variable dens2 equal density*density
variable temp equal temp
variable volume equal vol
variable temp2 equal temp*temp
variable press equal press
variable press2 equal press*press
variable N2 equal atoms*atoms
variable E2 equal etotal*etotal
variable E equal etotal
fix aves all ave/time 100 1000 100000 v_dens v_temp v_press v_dens2 v_temp2 v_press2 v_E v_E2
unfix nvt
fix nve all nve
run 100000
variable avedens equal f_aves[1]
variable avetemp equal f_aves[2]
variable avepress equal f_aves[3]
variable errdens equal sqrt(f_aves[4]-f_aves[1]*f_aves[1])
variable errtemp equal sqrt(f_aves[5]-f_aves[2]*f_aves[2])
variable errpress equal sqrt(f_aves[6]-f_aves[3]*f_aves[3])
variable heatcapacity equal ${N2}*(f_aves[8]-f_aves[7]*f_aves[7])/f_aves[5]
variable CV equal ${heatcapacity}/${volume}
print "Averages"
print "--------"
print "Density: ${avedens}"
print "Stderr: ${errdens}"
print "Temperature: ${avetemp}"
print "Stderr: ${errtemp}"
print "Pressure: ${avepress}"
print "Stderr: ${errpress}"
print "heatcapacity: ${heatcapacity}"
print "volume: ${volume}"
print "heatcapacity/volume: ${CV}"
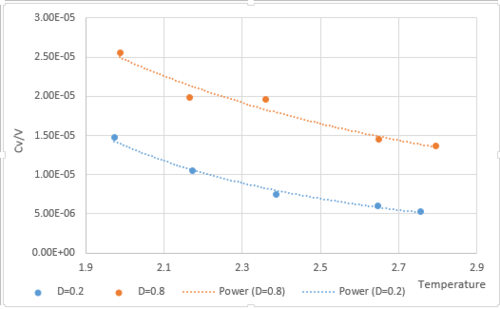
was plotted against temperature. The volume for is 16875. The volume for is 4218.75. The heat capacity is inversely proportional to temperature from Fig.8, the same as shown in the equation . This is because the lattice energy gap decreases with increasing temperature, so less energy will be required. This indicates that heat capacity is proportional to energy as shown in the equation. Also, it is shown that the lower the density, the lower the heat capacity.This is because high density means the particles will be closer together with lower volume, therefore less heat is required to heat the system. For the same number of particles, if the density is lower, that means the volume the particles take up is larger. Therefore the heat required is higher .
JC: The heat capacity is not necessarily inversely proportional to temperature because <E> and <E^2> are also dependent on temperature. You say that at higher density less heat is needed to raise the temperature which should mean the heat capacity is smaller, this is not what your results show.
Structural properties and the radial distribution function
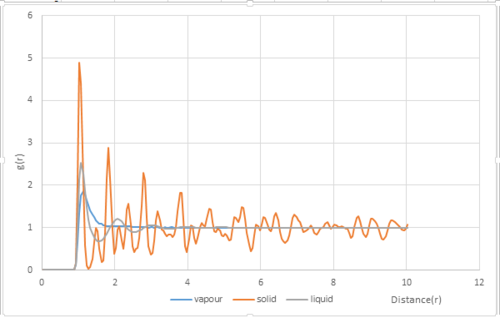
The radial distribution function was plotted for vapour, liquid and solid phases(Fig.9). The densities and temperatures were chosen from the phase diagram for the Lennard-Jones diagram.[3]:
| Phase | Density | Temperature |
|---|---|---|
| Vapour | 0.1 | 1.2 |
| Liquid | 0.8 | 1.2 |
| Solid | 1.6 | 1.2 |
The radial distribution function indicates the probability of finding a nearest neighbor from a particle. It will reveal the phase of the system simulated. The RDFs for the three systems are very different. The solid has the largest number of peaks followed by liquid and then gas. The peaks represent the density around each atom and hence solid which has the highest density will have more peaks. The peaks in the solid phase has decreasing amplitude with increasing r. It can be seen that the probability of finding a particle between the first and second peak is zero. This is because particles in solid phase do not have brownian motion and can only vibrate in fixed positions. The solid phase has long and short range order and this can be indicated by the peaks. The short range order is shown by the first three tall peaks and the long range order is shown by the smaller peaks behind.
JC: More peaks don't indicate a higher density, the RDFs are normalised - they all tend to 1 - and so you cannot tell which has a higher density. The RDF just indicates order.
The RDF of the liquid phase has three peaks with decreasing amplitude as r increases. The wider peaks mean that the liquid phase is more disordered than the solid phase.The decreasing amplitude with increasing interatomic separation indicates that the Brownian motion of particles in the liquid phase makes the order decrease with increasing separation. The three peaks indicate that the liquid phase only has short range order.
The RDF of the gas phase only has one broad peak. This suggests the gas phase is highly disordered and there is no short nor long range order.
In the RDF of the solid phase, the first three peaks correspond to the nearest neighbor of the referenced particle, the second nearest particle and the third nearest particle respectively. The lattice spacing is the distance between the zero probability minima and is 1.275 in reduced units.
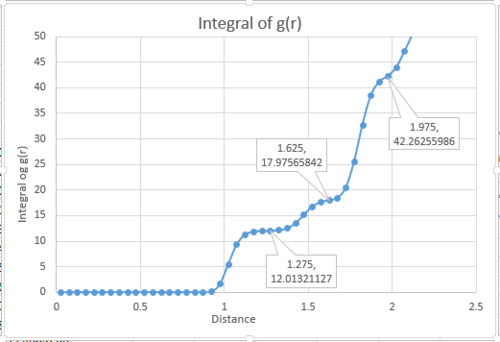
The coordination number for the first three peaks can be calculated from the plot of the integral of g(r) against interatomic distance. The integral of g(r) at the inflection points represent the coordination number of the three nearest neighbors. As a FCC lattice is used in a solid system, there should be 12 neighboring particles around each particle (shown at r=1.275). So the coordination number of the first peak is 12. The next inflection number has a g(r) integral of 18. As it is a running integral, the coordination number of the second peak is . The coordination number of the third peak is .
JC: It would have been good to show on a diagram of an fcc lattice which atoms are responsible for the first 3 peaks. Did you calculate the lattice parameter?
Dynamical properties and the diffusion coefficient
Mean Squared Displacement
The mean squared displacement is given by:
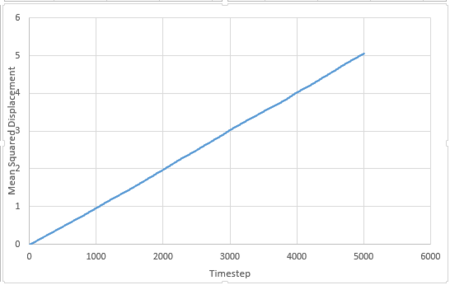
For 3375 atoms:
 |
 |
 |
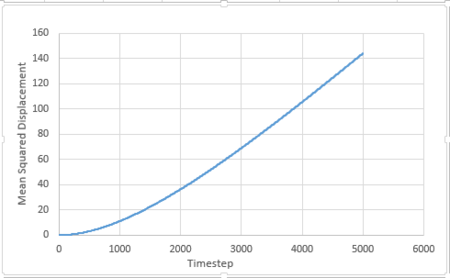
For 1 million atoms:
 |
 |
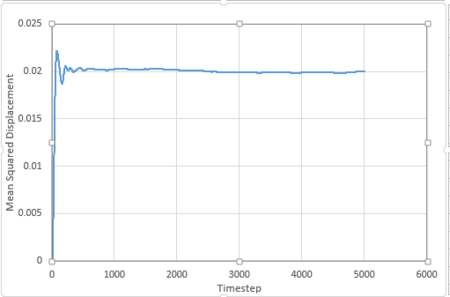 |
All these plots are as expected. For the liquid phase, MSD is directly proportional to timestep, this is because the particles move in brownian motion. Foe gaseous phase, the first part (timestep 0-2000) is curved and the second part (above 2000) is linear. The curved part is because the particles move randomly in the system and the distance between them is very large. The frequency of collision between the particles is very low and thus the velocity of the atoms will be almost constant. The distance travelled per unit time is constant, thus MSD is proportional to . As longer time is simulated, collisions will occur more frequently and the motion can be described by brownian motion and MSD changes linearly with timestep. For solid phase, the particles only vibrate in fixed positions and do not have enough kinetic energy to diffuse, thus MSD reaches at constant value at around timestep 200.
JC: The curved part of the gas plot is due to balistic motion, not because the particles move randomly. Show the lines of best fit on the graphs.
The diffusion coefficient is given by:
is the slope of the trendline of the mean squared displacement vs. timestep plot. The timestep .
For small system of 3375 atoms, the diffusion coefficient is: Liquid: ; Gas: ; Solid: ;
For large system of 1 million atoms, the diffusion coefficient is: Liquid: ; Gas: ; Solid: .
All of the diffusion coefficients are in reduced units. The coefficients for the larger system were similar to the ones for the smaller system.
Velocity Autocorrelation Function
The equation of the position of a 1D harmonic oscillator is:
, thus:
and
Therefore, by substitution:
Since :
Since :
As is an odd function, the area above the x-axis and below the x-axis cancel out from negative infinity to positive infinity. Thus, . therefore:
JC: Nice clear derivation and well spotted that you can simplify the integrals. cos*sin is also an odd function (even x odd = odd) so you don't need to change this to sin(2x).

The minima in the VACF for the liquid system represent the collisions between the atoms and the solvent molecules and change in direction. The minima in the VACF for the solid system represent the collisions between the atoms and change in direction. The minima for the solid system is lower than the minima for the liquid system because of the stronger interatomic forces. The VACF for the liquid system only has one weak oscillation, this is because the atoms only interact with their closest neighbor. In the VACF for the solid system, there are more oscillations as the atoms can vibrate in fixed positions. The harmonic oscillator VACF is very different to the Lennard Jones liquid and solid as there are no interactions between the atoms so the atoms will always vibrate with constant velocity without loss in energy. Therefore, the amplitude doe not change.
JC: There is not only one collision in the liquid, but the first collision randomises the particle velocities and the VACF decays to zero more quickly than in a solid.
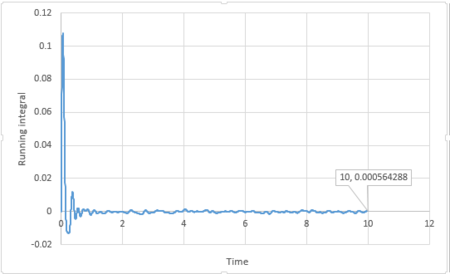
By applying the trapezium rule, integral under VACF can be calculated and running integral can be plotted:
For 3375 atoms:
 |
 |
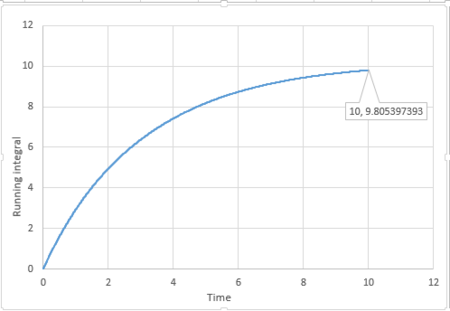
 |
For 1 million atoms:
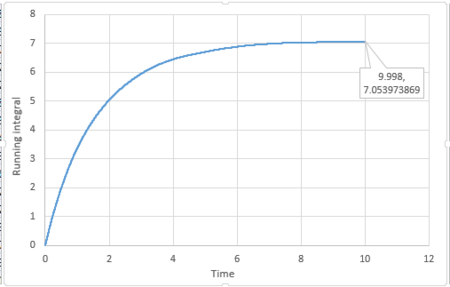
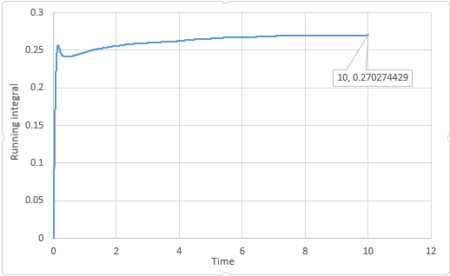 |
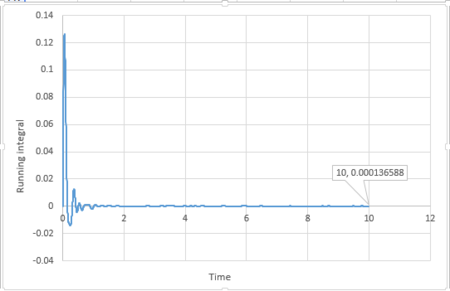 |
 |
The diffusion coefficient is calculated by:
The last point of the running integral is .
For 3375 atoms: Liquid: ; Solid: ; Gas: ;
For 1 million atoms: Liquid: ; Solid: ; Gas: .
The diffusion coefficient calculated from this method was largest for gas, followed by liquid and then gas. The coefficients for the larger system were very similar to the ones for the smaller system. The coefficients calculated by MSD were similar to the ones calculated by VACF for liquid and gas, but the coefficient calculated by VACF was larger than the one calculated by MSD for solid. The largest source of error may be that the trapezium rule overestimates the area under the solid curve as the timestep is not small enough.
JC: The trapezium rule is a source of error, but does it definitely overestimate the integral?
