Talk:Mod:fpp1994
Introduction to Molecular Dynamics
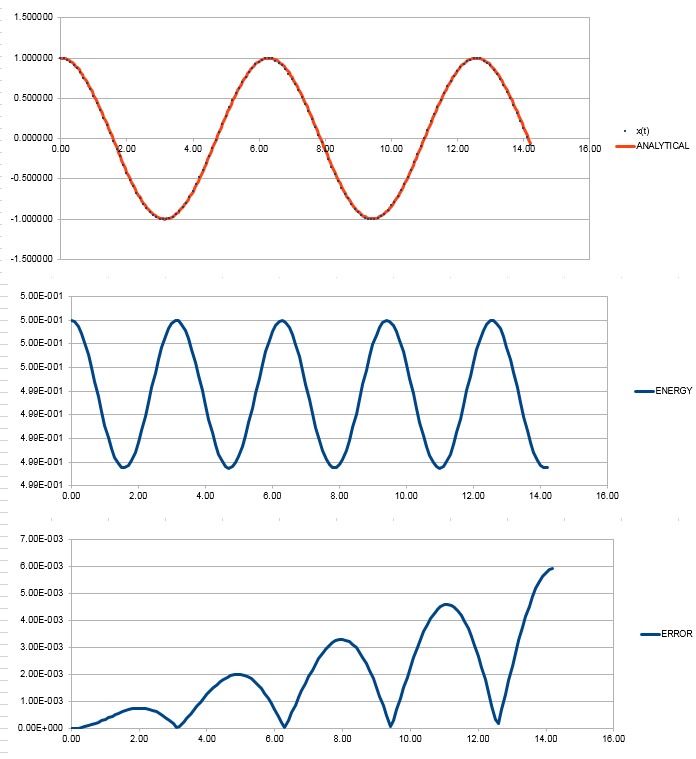
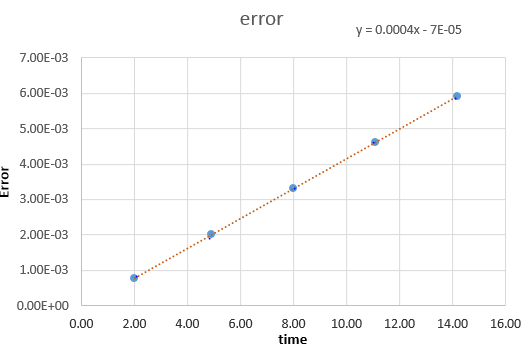
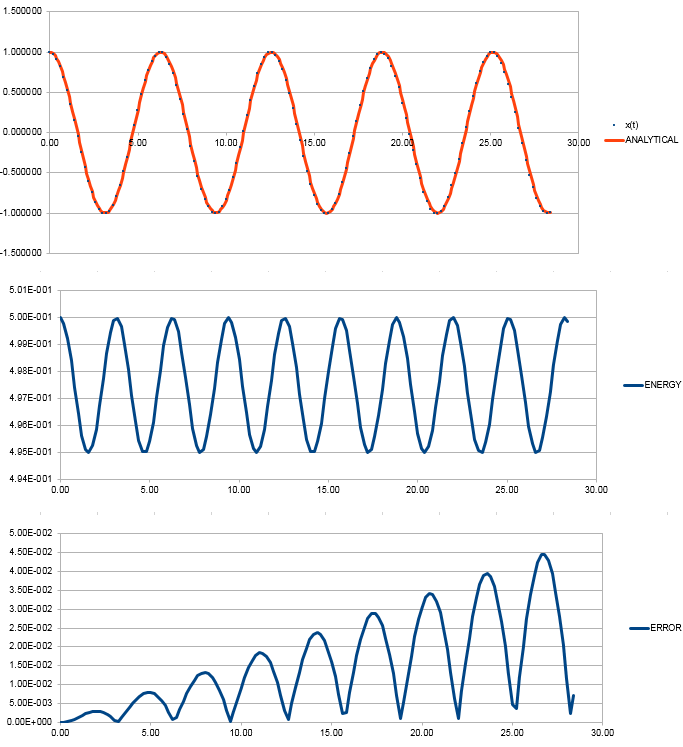
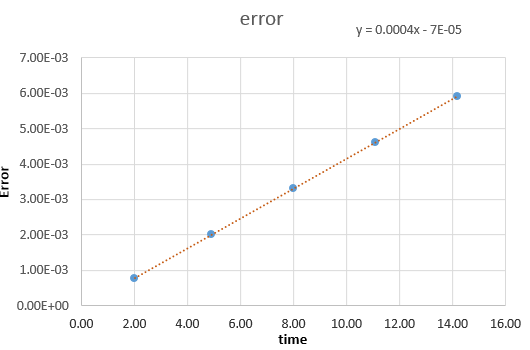
File:FppTASK3.xls Here, it can be seen that when the time-step is 0.2 the total energy fluctuates by 1%. It is important to model the energy of a system you are modelling because in some cases it can be used to calculate thermodynamic quantities as energy is what will dictate how things behave. NJ: It's more important to monitor the conservation of energy, this is an indicator of how well the numerical algorithm is modelling the system. NJ: Why do you think the error accumulates in the way that it does?
Task 4
- If
Thus ie is the distance when the potential is zero
- At this seperation, the force is
and
NJ: Be careful with the notation, , so this is out by a factor of -1.
- The equilibrium seperation, occurs at the bottom of the well, so force is at a minimum.
NJ: The potential is at a minimum, so the force is zero.
- and from just above
NJ: Good
So NJ: You should get a value of here
The expansion of the above bracket is but since and this becomes
Using the same expansion, but and as the lower limits gives and respectively.
NJ: Good
Task 5:
The density of water is and so would weigh . The molecular mass of water is and so the amount of molecules in of water is . Therefore molecules would occupy .
NJ: Good
Task 6
Consider an atom at position in a cubic simulation box which runs from to . In a single timestep, it moves along the vector . At what point does it end up, after the periodic boundary conditions have been applied?. It would be at
Task 7
- therefore
NJ: 1.088nm
- If then since the well depth in is
NJ: This is just in kJ, not kJ per mole.
- temperature therefore
Equilibration
Task 1 Giving atoms random starting positions in simulations can cause problems because if there are two atoms that are placed too close together (ie if they are within of each other) then their potential energies - and thus initial accelerations and velocities -will be extremely high and they will move through the sample with an unrealistically high speed and this will also disrupt other atoms.
NJ: Good. Can you say a bit more about this? Why won't this energy just be dissipated through the system?
Task 2
If each the spacing lattice point is 1.07722 and the lattice is three dimensional then the number density of lattice point is given by
If a face-centred cubic lattice has a number density of 1.2 then because it has four atoms per cell its volume can be given by and so the side length is just the cube root of this and so
Task 3
In the same lattice if the command "create_atoms" was used, 4000 atoms would be created as there are 4 atoms per unit cell.
NJ: Good. Nicely explained.
Task 4
mass 1 1.0 pair_style lj/cut 3.0 pair_coeff * * 1.0 1.0
The first command sets the relative mass of Atom 1 as 1. NJ: Good effort, but it's the mass of all atoms of type one, not just the mass of the first atom. In the second command "pair_style" is used to describe interactions between two particles and the "lj/cut" part tells LAMMPS that this interaction follows the Lennard-Jones potential and nothing more and that it cuts of when .
"pair_coeff" tells LAMMPS what coefficients to use in the style defined above. In this case it is the Lennard-Jones potential, and so it is telling LAMMPS to set and as 1. NJ: Good
Task 5 Specifying velocity and position as starting conditions means the Velocity Verlet Algorithm should be used.
Task 6
Specifying variables rather than numerical values useful is because you may need to use that value at various points throughout the script. Using numerical values all over the script would become particularly annoying if you were running lots of simulations with the same variable multiple times as you would have to look through the whole script to find each value but with the dollar command you can just change it once. NJ: This section makes sure that the same length of time is simulated no matter what timestep is chosen.
Task 7
The system reaches equilibrium after a time of about 0.5
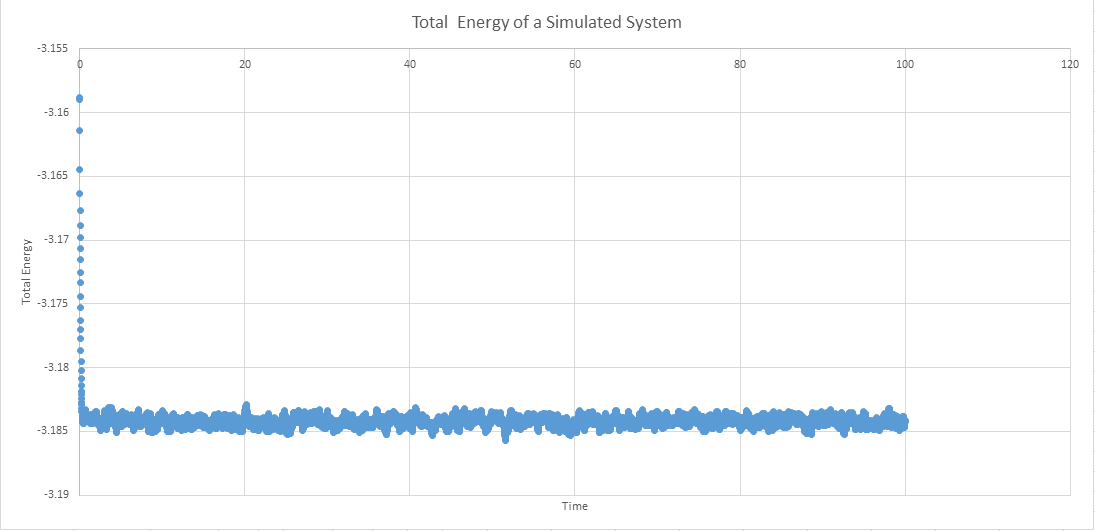
5 simulations were run with timestops of 0.001, 0.01, 0.0025, 0.0075, 0.015. The worst choice is the one with the highest timestop, 0.015 because ideally a balance between resolution and the amount of time the system can be monitored is preferred but all of the timestops show the eventual equilibrium and so there is little benefit to a large value as instead it does not show the fact that equilibrium had to be reached.
NJ: There should be a plot of energy as a function of time for all five timesteps, on the same axes. You should find that 0.015 does not reach equilibrium. What timestep did you choose for the rest of your simulations?
Running simulations under specific conditions
Task 1
10 phase points (two pressures and 5 temperatures) were selected, and and the timestep was 0.001
Task 2
therefore
Cancel to give
Task 3
From the script
fix aves all ave/time 100 1000 100000 v_dens v_temp v_press v_dens2 v_temp2 v_press2
From the LAMMPS manual
fix ID group-ID ave/time Nevery Nrepeat Nfreq value1 value2 ... keyword args ...
The numbers 100, 1000 and 100000 correspond to Nevery, Nrepeat and Nfreq respectively. These commands tell LAMMPS on which timesteps to calculate the averages of the properties that follow. Nrepeat tells LAMMPS how many averages to calculate, starting from the value given by Nfreq and working back in multiples of Nevery. In this case 1000 averages are taken in total, once every 100 timesteps up to 100000 which is as far as the script will run.
Task 4
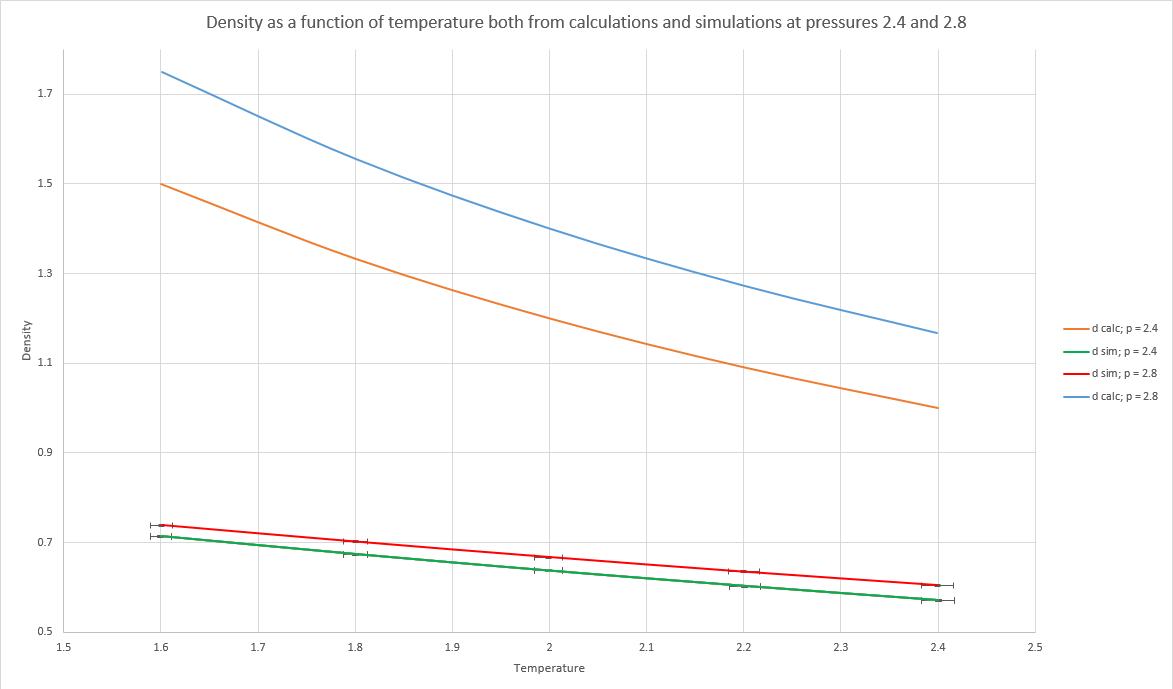
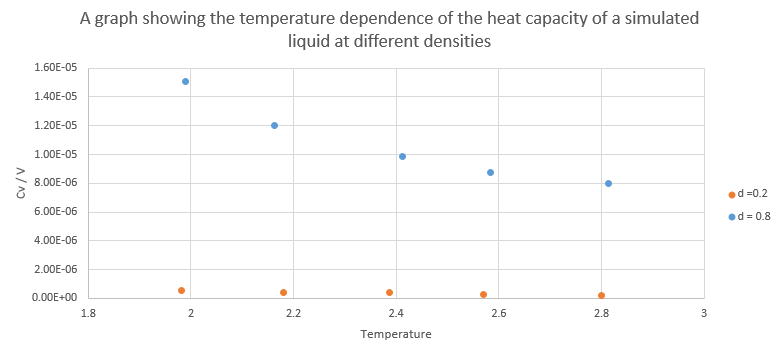
The bottom two lines are the density as a function of temperature as calculated by LAMMPS whereas the top lines are calculated using the ideal gas law using the equation derived below where pressure was 2.4 and 2.8 and the temperature was the LAMMPS average. The simulated density is much lower than the one predicted by the gas law because the gas is not ideal as there are repulsive and attractive terms in the Lennard-Jones potential and ideal gases have no interaction between the particles. NJ: Good, but why do you always find a lower density in the simulations?And as the pressure increases the gap between the simulated and calculated densities increases too because at higher pressures there are more interaction between the particles and so the ideal gas approximation gets worse. NJ: Good. What about the trend with temperature?
and
so but on LAMMPS in lj style, and are all equal to 1
so
Calculating heat capacities using statistical physics
The script used to calculate the heat capacities is below
### DEFINE SIMULATION BOX GEOMETRY ###
variable density equal 0.2
lattice sc ${density}
region box block 0 15 0 15 0 15
create_box 1 box
create_atoms 1 box
variable atoms equal 15*15*15*${density}
### DEFINE PHYSICAL PROPERTIES OF ATOMS ###
mass 1 1.0
pair_style lj/cut/opt 3.0
pair_coeff 1 1 1.0 1.0
neighbor 2.0 bin
### SPECIFY THE REQUIRED THERMODYNAMIC STATE ###
variable T equal 2.0
variable timestep equal 0.001
### ASSIGN ATOMIC VELOCITIES ###
velocity all create ${T} 12345 dist gaussian rot yes mom yes
### SPECIFY ENSEMBLE ###
timestep ${timestep}
fix nve all nve
### THERMODYNAMIC OUTPUT CONTROL ###
thermo_style custom time etotal temp press
thermo 10
### RECORD TRAJECTORY ###
dump traj all custom 1000 output-1 id x y z
### SPECIFY TIMESTEP ###
### RUN SIMULATION TO MELT CRYSTAL ###
run 10000
unfix nve
reset_timestep 0
### BRING SYSTEM TO REQUIRED STATE ###
variable tdamp equal ${timestep}*100
variable pdamp equal ${timestep}*1000
fix nvt all nvt temp ${T} ${T} ${tdamp}
run 10000
reset_timestep 0
unfix nvt
fix nve all nve
### MEASURE SYSTEM STATE ###
thermo_style custom step etotal temp press density
variable dens equal density
variable dens2 equal density*density
variable temp equal temp
variable temp2 equal temp*temp
variable press equal press
variable press2 equal press*press
variable etotal equal etotal
variable etotal2 equal etotal*etotal
fix aves all ave/time 100 1000 100000 v_dens v_temp v_press v_dens2 v_temp2 v_press2 v_etotal v_etotal2
run 100000
variable avedens equal f_aves[1]
variable avetemp equal f_aves[2]
variable avepress equal f_aves[3]
variable aveetotal equal f_aves[7]
variable aveetotal2 equal f_aves[8]
variable errdens equal sqrt(f_aves[4]-f_aves[1]*f_aves[1])
variable errtemp equal sqrt(f_aves[5]-f_aves[2]*f_aves[2])
variable errpress equal sqrt(f_aves[6]-f_aves[3]*f_aves[3])
variable heatcap equal ${atoms}*${atoms}*((f_aves[8]-f_aves[7]*f_aves[7])/(f_aves[2]*f_aves[2]))
print "Averages"
print "--------"
print "Density: ${avedens}"
print "Stderr: ${errdens}"
print "Temperature: ${avetemp}"
print "Stderr: ${errtemp}"
print "Pressure: ${avepress}"
print "Stderr: ${errpress}"
print "Heat capacity: ${heatcap}"
Structural properties and the radial distribution function
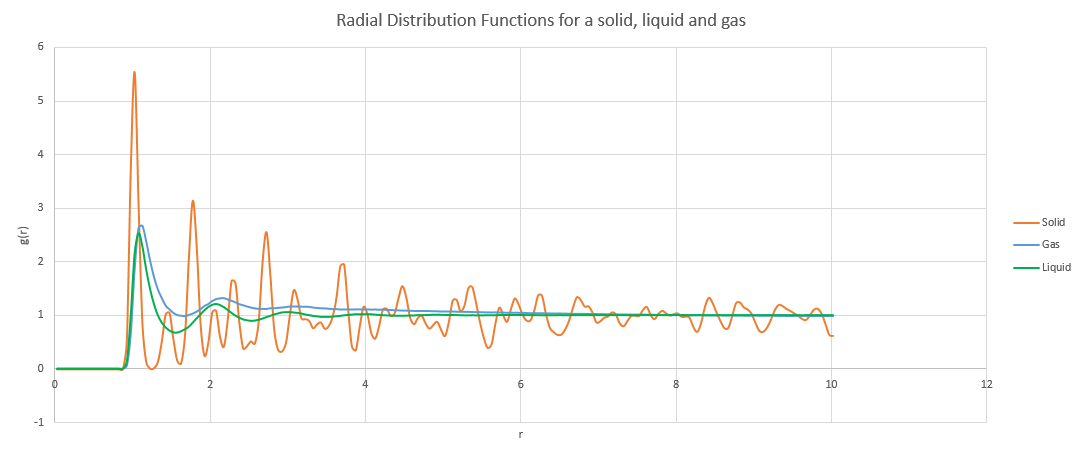
NJ: I think there's a little bit of confusion here - whichever reference atom you pick, the lattice looks the same in all directions. The RDF is also averaged over all atoms. I wanted you to work out the interatomic distances in terms of the lattice constant for the first three peaks, and calculate the spacing using your graph, rather than reading from the log file. You also should have used the integrated g(r) data to calculate the coordination number for each of the first three peaks.
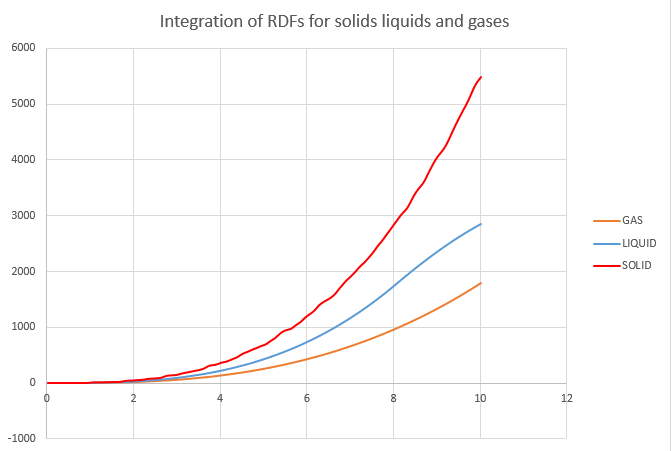
The RDF for liquids and gases look quite similar to each other but different to that of the solid as they are much less ordered. In both cases, the first peak is quite large before the curve quickly levels out at 1. The RDF measures the amount of atoms in a shell of radius, r, around the reference atom and so when the shell is small the concentration of nearby atoms is high and they resemble clusters. However, as the shell gets bigger it starts to become more diffuse and eventually reaches a point where the concentration of atoms in the shell cannot be distinguished from the density of the system as a whole.
NJ: What density did you use for the gas? As you say, it looks very similar to the liquid. I suspect this is a bit too dense, and is in the liquid/gas coexistence region. Why does the solid RDF have so many more defined peaks?
By eye, the liquid and gas RDFs would seem to suggest that the gas is ever so slightly denser than the liquid, however, the functions are normalised and the actual density of gases is much lower and so there are far fewer atoms than in the liquid, as can be seen in the plots of their integrations.
From the log file, the lattice spacing of the solid is
Lattice spacing in x,y,z = 1.45447 1.45447 1.45447
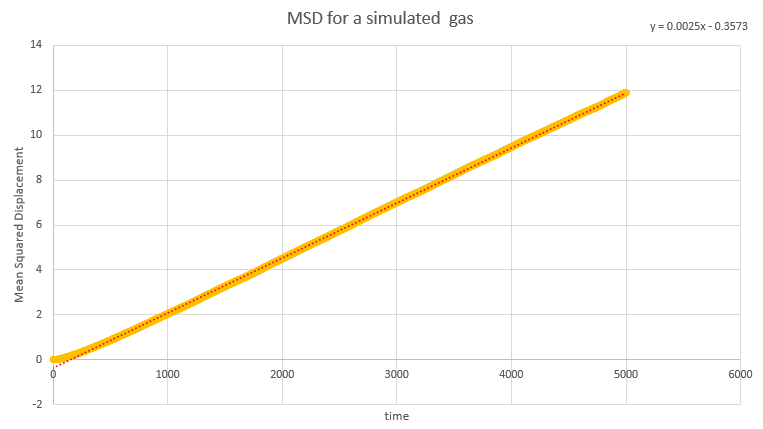
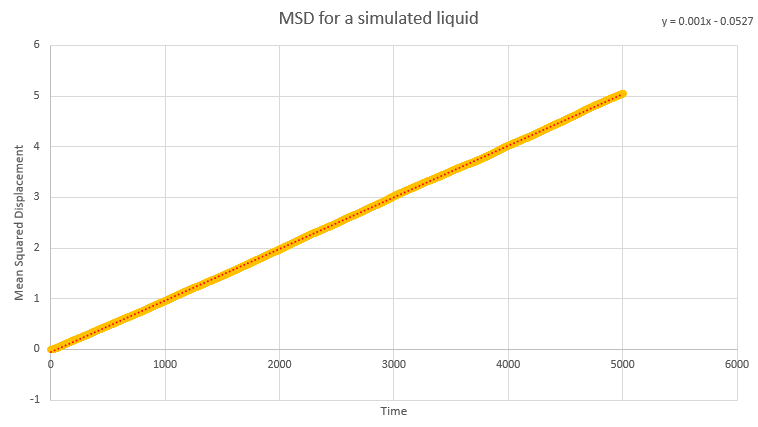
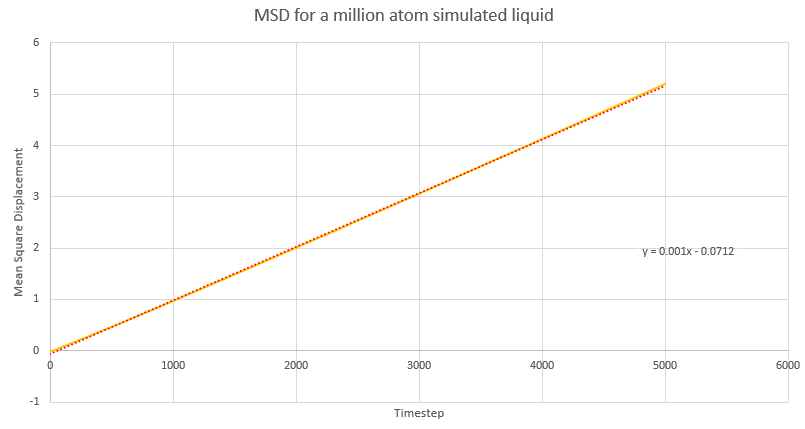
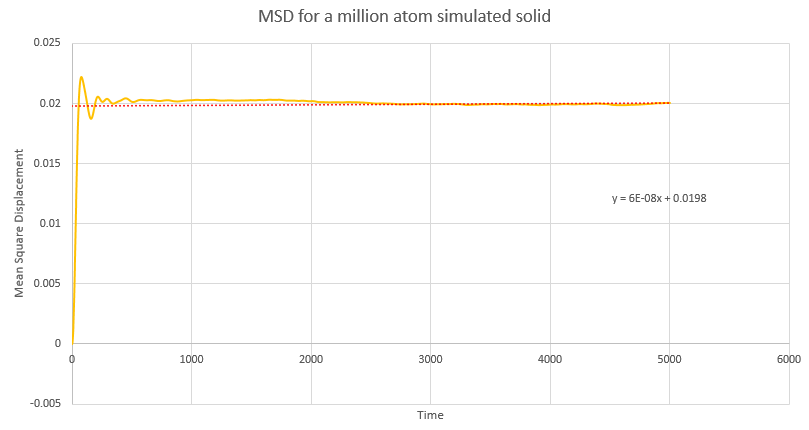
Dynamical properties and the diffusion coefficient
NJ: Be careful, these values are all in reduced units.

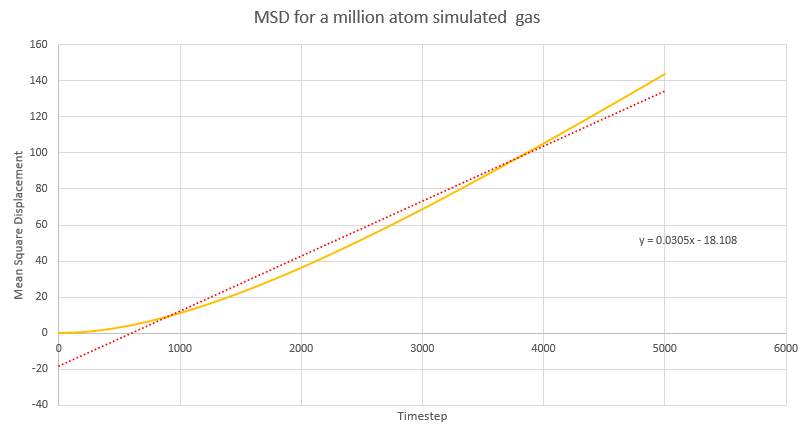
NJ: How do the values of D compare across phases? Does using 1 million atoms make a difference?
define:
NJ: This is the right start.