Talk:Mod:Hunt Research Group/How to draw SDFs with TRAVIS
TRAVIS is a free tool for analyzing and visualizing trajectories from MD simulations.
TRAVIS source code can be obtained for free from "Source Code (.tar.gz)" at Download page on TRAVIS website.[1] How to compile and Install TRAVIS is explained on Page 3 in "Quick Start Guide (PDF)" which can be downloaded on the same link.
Details about TRAVIS is explained in the paper.[2]
M. Brehm and B. Kirchner. J. Chem. Inf. Model. 2011, 51 (8), pp 2007-2023.
Manual in Word format is obtained here. How to generate SDF.docx
- [N1444][NTf2] HISTORY(trajectory) file for trial use is here. Media:HISTORY.txt
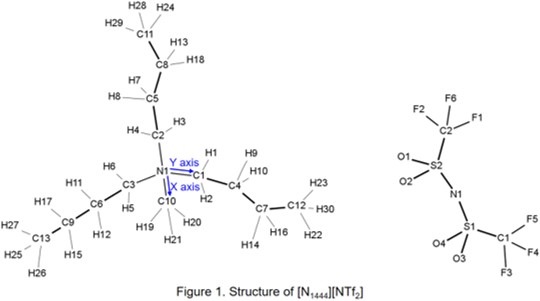
- NB: When you use this file with TRAVIS, change the file name from HISTORY.txt to HISTORY before using. [N1444][NTf2]= tri-n-butylmethylammonium bis-(trifluoromethanesulfonyl)imide (Fig. 1)
How to draw SDFs with TRAVIS
- The explanation below is written to explain how to draw spatial distribution of anions in the first shell around a cation by using [N1444][NTf2] as an example.
1. Create a new directory and put HISTORY file there.
- HISTORY file is so huge that it takes much time to copy. It is recommended to make a link from the original location by executing “ln -s (Original location)/HISTORY HISTORY” in the created directory.
2. Execute “travis HISTORY” in the created directory, and answer the displayed questions based on your model (and preference). Default answers are shown in brackets and when you choose the default answers, just type Enter. Questions and answers shown below are based on [N1444][NTf2].
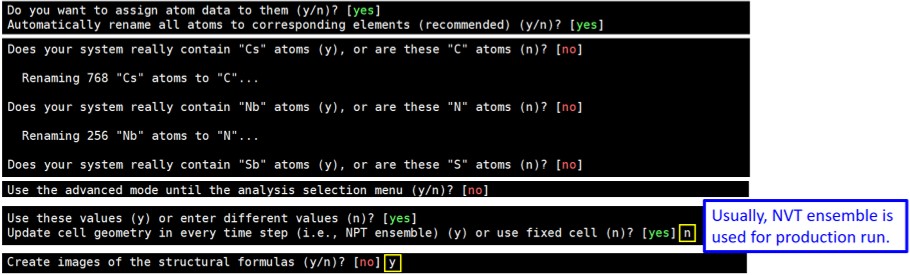
After answering “Create images of the structural formulas?”, press “Ctrl” and “c” at the same to quit once.
Then, execute “ls -l”, and you will find “.dot” files.
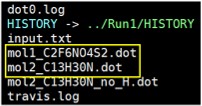
3. Visualise the “.dot” files. Choose from the options below.
3-1. Using Graphviz (Recommended) Visit https:/www.graphviz.org/download/ and download Graphviz.
Then, execute “neato -Tpng mol1_****.dot -o mol1_***.png”.
In [N1444][NTf2] example, execute “neato -Tpng mol1_C2F6NO4S2.dot -o mol1_C2F6NO4S2.png”.
Likewise, execute “neato -Tpng mol2_****.dot -o mol2_***.png”.
In [N1444][NTf2] example, execute “neato -Tpng mol1_C13H30N.dot -o mol1_C13H30N.png”.
When you execute “ls -l”, you will find two png files. After sending these png files to your local machine, open them and confirm the chemical structures.
In [N1444][NTf2] example, you will find the structure shown in Fig 1.
3-2. Handwriting the chemical structure (Recommended only if the structure is simple)
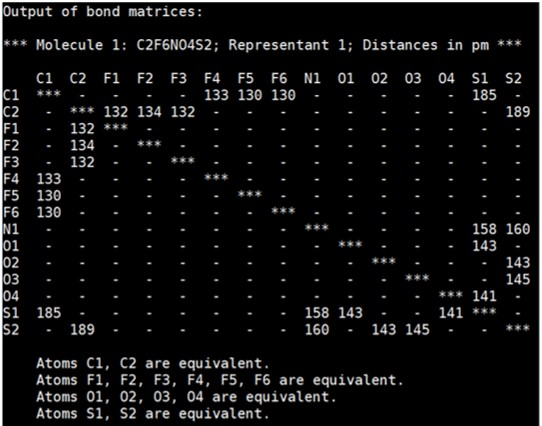
In the screen, you will find “bond matrices” shown as below. Thus, you can handwrite the chemical structure based on this information.
For example, the matrices below mean C1 is bonded to F4, F5, F6, and S1.
4. Return to the directory created in Step 1, and execute “travis HISTORY” again. Restart answering the questions. When you face “Create images of the structural formulas?”, answer no (just type Enter) this time, and keep answering until the end.
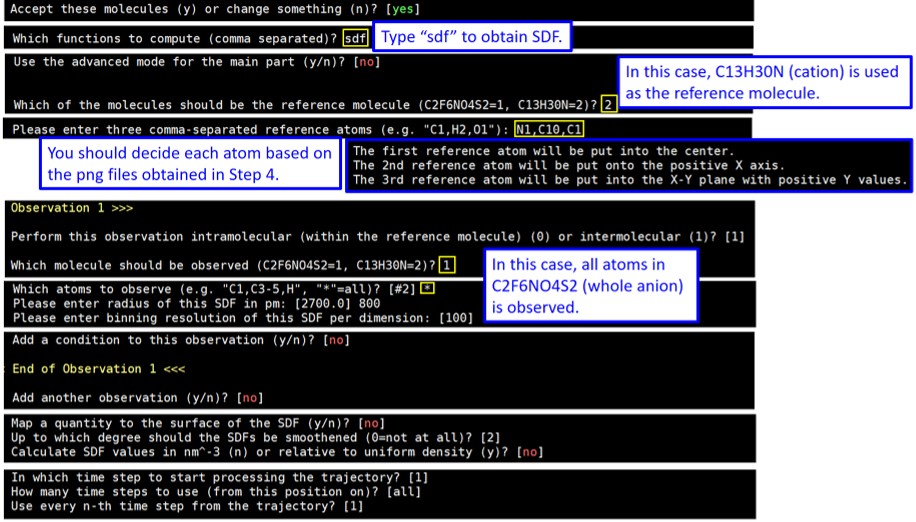
5. When you finish answering all questions, TRAVIS starts creating SDF files. W ait until ***The End*** is shown. Then, execute “ls -l”, and you will find the created files. Send SDF files(.plt) and ref file to your local machine.

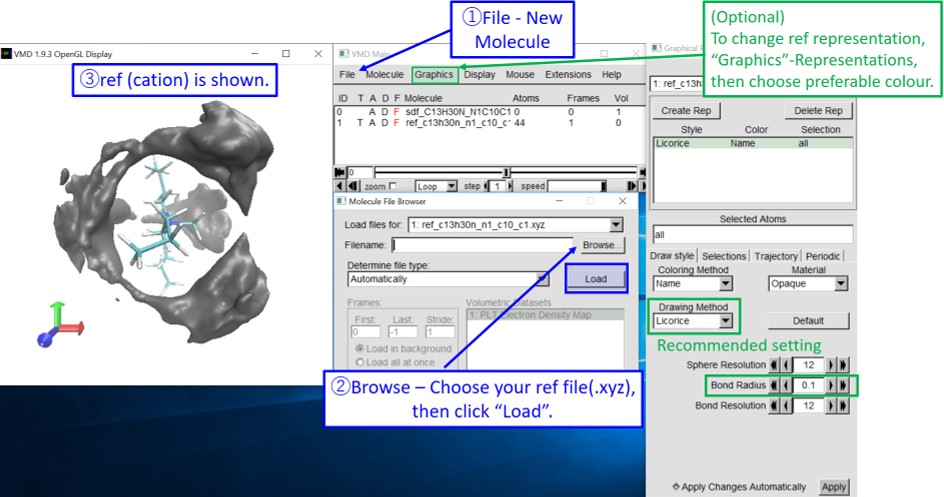
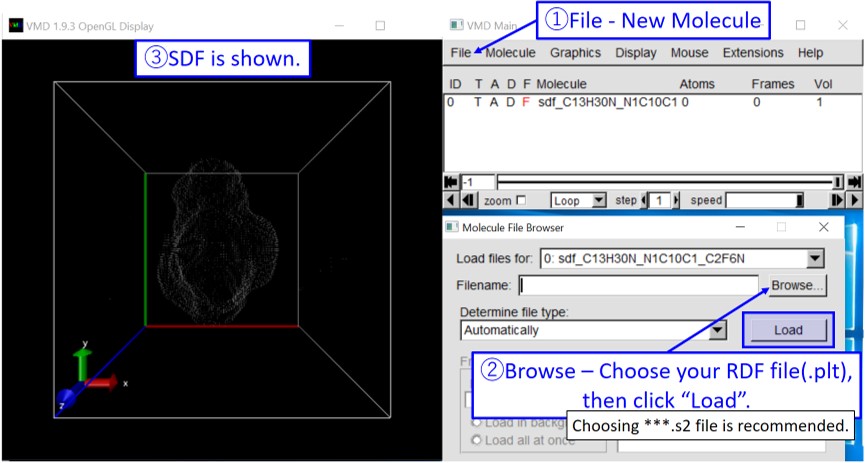
6. Start VMD. Open your SDF by following the instruction below.
(Options)
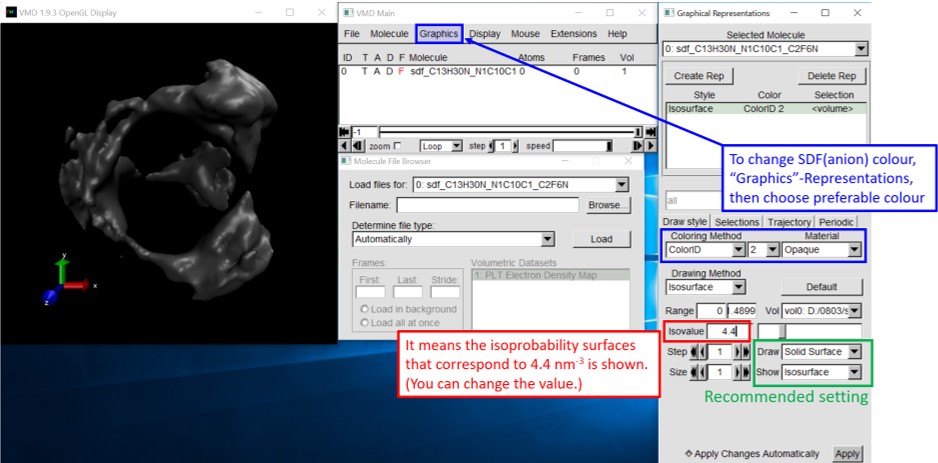
6-1. Change the SDF graphics based on your preference.
6-2. Change the background colour based on your preference.
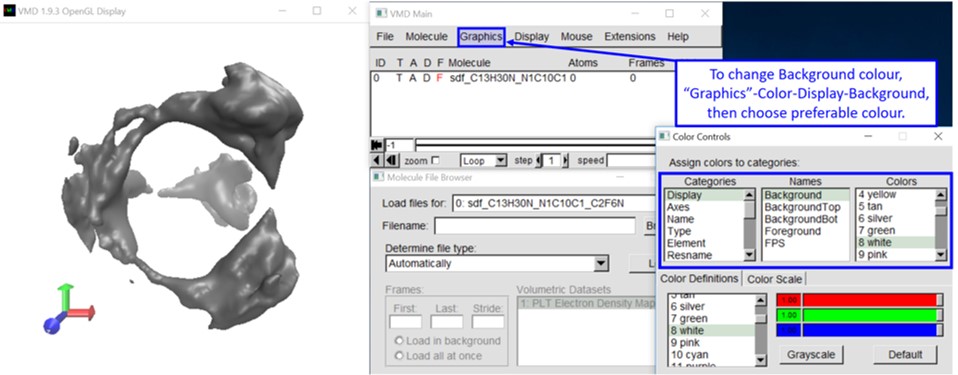
6-3. Change the input file based on your preference.

7. Insert the reference molecule (cation) by following the instruction below.