Talk:Mod:Hunt Research Group/Calc XPS Code
Links to Files
Open Shell Version: Media:Gelius_open_shell_code_Aug2016.zip
Closed Shell Version: Media:Gelius_closed_shell_code_Aug2016.zip
Plot Atomic Contribs Script: Media:Plot Atomic Contribs V3.m
- MLinpt.txt File parser(Matlab): Media:ML inpt att2.m
Introduction
The purpose of this code is to calculate partial density of states (pDoS) plots and Valence band x-ray photoelectron (XP) spectra from .fchk files. There is both an open shell and a closed shell version of the code, they work in exactly the same way.
All code was written using python 2.7 (with numpy), Multiwfn version 3.3.8, Matlab version 2014a and a cygwin(bash) shell on windows 8. It should work on windows or linux if the correct options are set. It might not work on a mac currently due to Multiwfn not working properly (I haven't personally tested it though).
I've now also uploaded a parser for *MLinpt.txt files. Documentation is in the file itself, this is a totally non-essential code but can make writing scripts that rely on *MLinpt.txt files easier.(e.g scripts to extract HOMO energies).
Some Theory
Mulliken Population Analysis
A pretty key part to this code is the breakdown of molecular orbitals (MOs) in atomic orbitals (AOs, which are essentially basis functions). This is carried out in multiwfn using Mulliken population analysis, which i'll briefly summarise due to its importance.
The basic question is "For MO X, how many electrons come from atomic orbital Y?". Note im going to be using "basis functions" instead of "atomic orbitals" from this point, in terms of DFT calculations we interpret a basis function centered on atom X as an atomic orbital of atom X.We'll define as the wavefunction for a single MO containing a single electron. Next we write as the linear combination of individual basis functions ():
Since we want a number of electrons, we're interested in the formula for the electron density () of this MO:
If these formulae are tricky to follow, try doing it for a system with just two basis functions. Note that in the third line the indice j runs over the same coefficients and basis functions as the indice i, its just used to show that the term only contains products between 2 different basis functions/co-efficients. Hence, we now have a formula for the MO density which is purely in terms of (known) basis functions and (known) coefficients. Now its just a case of working out how we should interpret these values.
Firstly note that (since we use normalised basis functions) hence:
The values of c_{i} in this expression are easy to interpret, they represent the fractional contribution of AO i to the wavefunction. The term is more difficult to interpret, each expression in the series can be thought of as the number of electrons shared between two basis functions. i.e. the following term would represent the number of electrons shared between basis function 1 and 2:
However, we need to split this "shared" electron density between the two contributing basis functions. In Mulliken Population analysis "shared" electron density is divided equally between the two contributing basis functions. So the total electron density assigned to one basis function can be written as:
represents the fractional contribution of i to the wavefunction. Hence if you had a 2-electron MO, you'd simply multiply by 2.
What the code actually does (roughly/briefly)
Step 1 - The first step is to run a Mulliken population analysis on the *.fchk file. This leads to a file containing the energy/AO composition of each MO in the system (the *MLinpt.txt file). Subsequent steps vary depending on if you want partial density of states (pDoS) plots or calculated valence spectra.
If calculating pDoS (not XPS spectra):
For each type of atomic orbital present do the following:
- For each MO, center a gaussian-lorentzian function at that MO energy. Multiply the intensity by the contribution of the current AO to the current MO.
- Sum all the resultant peaks, this generates the pDOS the current atomic orbital
Once all pDoS are generated, the total DoS could be created by summing them all. In practice the code probably just calculates the total DoS first, by just placing a gaussian-lorenztian with area=1 around each MO energy.
If calculating XPS spectra:
For each type of atomic orbital present do the following:
- Multiply the fractional contributions of this AO to each MO by the AO cross-section. This value will be reffered to as the "intensity" This cross-section depends on both the AO type and the photon energy. Default values in this code are stored in tables in X_Section_Library, and custom/different values can easily be added.
- For each MO, center a gaussian-lorentzian function at that MO energy. This area of this function is equal to the intensity calculated in the step above.
Sum all the resultants peaks, this generates the contribution of the current AO to the valence XPS spectrum Once this is repeated for each AO type, sum all those individual contributions to generate the total XPS valence spectrum.
"Installation" Instructions
All instructions will be given for the open shell version of the code. However, the process is the same except whenever these instructions say to edit the "gen_gelius_input_opensh.sh", just do the corresponding edit on the gen_gelius_input.sh file. The Matlab scripts are EXACTLY the same for both open shell/closed shell codes.
0) Make sure you have multiwfn (http://sobereva.com/multiwfn/) saved somewhere, python 2.x and numpy installed.
- a)On ubuntu Multiwfn needs an extra library, type the following line:
sudo apt-get install libmotif-dev
1)Put the following files (all the .py/.sh files) somewhere in system path (e.g i use /usr/local/bin):
- a)Format_Multiwfn_Output_alpha.py (Format_Multiwfn_Output.py for closed shell)
- b)Format_Multilwfn_Output_beta.py
- c)Gen_Multiwfn_Input_Comms.py
- d)Gen_Multiwfn_Input_Comms_beta.py
- e)Merge_Fragment_Files.py
- f)gen_gelius_input_opensh.sh
Make sure all files (including Multiwfn) have executable persmission (navigate to the folder and do "chmod u+x filename")
2)In the gen_gelius_input_opensh.sh change the current lines accordingly:
- a) multiwfn_folder = 'C:/Users/Richard/Documents/Multiwfn_Installed' to the directory where you store multiwfn and associated files.
- b) "cygwin=1” to “cygwin=0” if your not using a Cygwin shell.
- c) “old_sed=1” to “old_sed=0” if your version of sed allows the –i option (edit in place) to be used. I don’t think the standard version on mac does(meaning set old_sed=1),standard version of Cygwin does though.
- d) If on linux "cp $multiwfn_folder/Multiwfn.exe $curr_folder" to "cp $multiwfn_folder/Multiwfn $curr_folder" . Do the same for the line where multiwfn is deleted from current folder("rm Multiwfn.exe").
3)Put the following files somewhere in matlabs path (e.g your matlab folder in documents)
- a) Get_X_Section.m
- b) Gen_Gelius_Spectrum.m
The Get_X_Section REALLY does need to be in matlabs path (unless you want to edit some code in Gen_Gelius_Spectrum.m). If you know what your doing in matlab feel free to put Gen_Gelius_Spectrum.m wherever you want.
4)Put the X_Section_Library folder wherever you want.
5) In Get_X_Section.m modify the following line:
- "Lib_Path ='C:\Users\Richard\Documents\X_Section_Library' " to the path where you put the X_Section_Library folder in.
6) OPTIONAL: Add the code in vim_template_script to your vimrc (c+p it somewhere in the file). "vim ~/.vimrc" to access your vimrc. You can then bring up a template input file in vim by using ":call Gelius_Input()" Since i forgot to put the relevant text file in to the code folders, just c+p the below into your vimrc to get this to work.
function! Gelius_Input()
exe 'normal aCoSCN4_freq_AllNCS_B3LYP_6311plgdp_Tetrahed_BMIMSCNSMD_Abr.fchk\rNumb_Fragments=10\r1a=\r1t=a\r1l=TDOS\r2a=\r2t=\r2l=\r3a=\r3t=\r3l=\r4a=\r4t=\r4l=\r5a=\r5t=\r5l=\r6a=\r6t=\r6l=\r7a=\r7t=\r7l=\r8a=\r8t=\r8l=\r9a=\r9t=\r9l=\r10a=\r10t=\r10l=\rM_factor=1.0\rLink_File='
exe '%s/\\r/\r/g'
endfunction
Running The Code
Using the Example files
1) Navigate (in your shell) to the folder "Example_files". e.g for me i ran:
"cd /cygdrive/c/Users/Richard/Documents/Random_Coding/Gelius_Spectrum_Automated_Jan_2016/open_shell_code/Example_files"
2) Run this command in console(for closed shell use gen_gelius_input.sh):
"gen_gelius_input_opensh.sh"
Command 2) will have created a folder for each .fchk file in Example_files/. Therefore you should now have folders:
- CoSCN4_freq_AllNCS_B3LYP_6311plgdp_Tetrahed_BMIMSCNSMD_Abr
- other_file
Within these folders are tons of output files. The most important ones are those ending in "MLinpt.txt" - these will be the input files for step 3.
The script will run on all .fchk files in the current folder, when you do this for real you need to make sure that all the fchks are for the same system (e.g a water molecule) and that the atomic numbering is the same in all files (view-->labels in gaussview gives you what im calling atomic numbering).
If anything went wrong try revisiting the installation instructions.
3) Everything from now on is done using matlab. Theres a file called "pathname_file" in the Example_files folder which was created by the gen_gelius_input_opensh.sh script. Copy its full contents to clipboard and in MATLAB terminal type:
plist=<CTRL-V> (<CTRL-V> means paste, rather than type out CTRL-V)
In matlab terms, plist now contains a cell array where each entry is the full path to a folder. The folder paths are those for the folders created in step 2). Hence to do this manually for just one of the two folders you'd type something like:
plist={'C:\Users\Richard\Documents\Random_Coding\Gelius_Spectrum_Automated_Jan_2016\open_shell_code\Example_files\other_file'}
4) To create either an XPS spectrum or pDoS plots we use the Gen_Gelius_Spectrum script. This takes the following three arguments IN THE FOLLOWING ORDER:
- a) the plist variable we just made
- b) the photon energy we want to use (any negative number requests a pDoS rather than an XPS spectrum)
- c) the FWHM of the peaks to place around each MO energy (units = eV)
So to create a pDoS plot you could type the following into the matlab terminal(Try this one first):
Gen_Gelius_Spectrum(plist,-1,1)
To create an XPS spectrum taken at photon energy of 1486eV (standard lab-based value) type the following:
Gen_Gelius_Spectrum(plist,1486,1)
If you get an error like "Undefined function 'Gen_Gelius_Spectrum' for input arguments of type 'cell'.", then your Gen_Gelius_Spectrum script probably isnt in your matlab path. (see installation instructions, or google a bit)
If your error free then well done. Sub-folders should have been created in "other_file" and "CoSCN4_freq_AllNCS_B3LYP_6311plgdp_Tetrahed_BMIMSCNSMD_Abr", which in turn will contain a loada output files. See the output files section of the wiki for an explanation of what these are.
Creating your own input files
It can be named anything (except a substring of the fchk file name). The extension must be .test.
If you followed the optional part above and made a vimscript, then a template for the input file can be brought up in vim by typing ":call Gelius_Input()"
An example input file is shown below. Each "fragment" (e.g cation,anion, alkyl group etc.) should have its own input file.
other_file.fchk Numb_Fragments=4 1a=10-13 1t=a 1l=TDOS 2a=10-13 2t=s 2l=S3s 3a=10-13 3t=p 3l=S3p 4a=10-13 4t=d+ 4l=S3d M_factor=1.0 Link_File=all_frags.test
The input file can be divided into parts as follows:
1) The top line of the input file tells the code the name of the checkpoint file to use in the calculation. In practice you can write ANYTHING on this top line if using the gen_gelius_input*.sh script, as the script itself replaces the first line (whatevers there) with the names of all .fchks in the current folder. But SOMETHING must be written there.
2)Numb_Fragments is the number of sub-fragments (max 10) that you want to break your pDoS down into (e.g fragment 2 corresponds to all Sulfur 3s atomic orbitals in the example above). Code will break if you get this wrong.
3) 1a/1t/1l instruct the code what to use as the total density of states. Hence 1a must be set to ALL the atoms your interested in for this fragment, 1t must be set to "a" and 1l should be set to TDOS (not sure it matters what this is set to, but to be on the safe side just use TDOS). The purpose of this fragment is essentially error checking.
4) 2a/3a/*a : a stands for atoms here, so the value of a corresponds to the labels of the atoms you want to be included in the sub-fragment. These labels are the same integers you get in gaussview when you go to view-->labels. Examples of valid options may be "1,4,5-7" or "1,3,5" (basically comma seperated list of integers)
5) 2t/3t/*t: t stands for "type of AO".This is a comma seperated list with possible arguments (any combination) "a,s,p,d,f,g". "a" corresponds to all types of AO centered of the atoms specified with the 2a/3a/*a line. s means s-type atomc orbitals (0 angular momentum), p means p-type atomic orbitals (1 angular momentum) etc. To get all AOs of a certain angular momentum and higher (up to g-type AOs) add a plus to the end of the relevant letter. e.g "p+" says "include all AOs with angular momemtnum greater than or equal to 1 in this sub-fragment") DON'T use "a" with any other letter, and dont do anything else that will lead to a type of AO being requested twice (e.g p+,d)
6) 2l/3l/*l: l stands for label. If you dont care about calculating an XPS spectrum you can essentially set it to whatever you want. This tells the matlab script what cross section to use for calculating the XPS spectrum, hence it must match (case insensitive) a filename in the X_Section_Library folder. e.g S3s tells matlab to get the S3S.txt file when looking for what cross-section to use.
7) M_factor=1.0, you really never need to modify this but make sure its present. It's purpose is to tell matlab to multiple the pDoS of this fragment (that this input file corresponds to) by the value M_factor. The purpose is for systems where the real stoichiometry differs from that in your calculation. For example if i have a mixture of 99 water:1 anion but only a calculation on 1 water molecule mixed with 1 anion i could set M_factor for the water fragment input file to M_factor=99.0 to estimate (badly) what the actual DoS of the mixture would look like.
8) Link_File=*. This options for if you have a fragment (e.g an anion) but want it broken into more than 10 sub-fragments (e.g this will be true for any molecule with >3 elements). In this case make 2 files for that fragment, and in the second file set Link_File to the name of the first file. In the example above, this input file will essentially be merged with "all_frags.test", and only an output file for all_frags.test will be generated.
More complex input file options
Gen_Gelius_Spectrum.m
This code has the following options that can be changed (all are explained by comments in the .m file):
1)min_e/max_e
Matlab ignores any orbitals with energy below min_e or above max_e when generating the XPS spectrum. Note if you want to change this to include core orbitals or very high energy virtual orbitals then you'll also have to edit Gen_Multiwfn_Input_Comms.py to set the range in multiwfn to the one you want.(Theres currently no variable in that .py code to do that, it just uses the defaults in multiwfn. But the function you'll need to edit in Gen_Multiwfn_Input_Comms.py is Output_Header.
2)min_e_plotted/max_e_plotted
These refer to the minimum/maximum binding energies to calculate spectral intensity for. The range should probably be slightly wider than the min_e/max_e range really, though thats not the default setting.
3)step_size
The value (in eV) between calculated data points in the output spectra. For example, if set to 0.001eV then a data point will be calculated for 0eV,0.001eV,0.002eV etc. Lower values=larger files/slower runtimes.
4)lorentz_fraction
Value between 0 and 1 that determines the % Lorentzian character of the lineshape used in the broadening function(the remaining % is set to gaussian). By broadening function i essentially mean the peak thats placed around every MO energy in generating these spectra. lorentz_fraction=0 will give you a pure gaussian peak, while lorentz_fraction=1 gives a pure Lorentzian peak.
5)savetext/figsave/savejpeg/all_contribs_output
These all control whether a certain type of output file is saved, with variable names that should be self-explanatory.Values of 1 mean save the file, other values mean dont save. set savetext=0 to not save any .txt output files, figsave=0 to not save any .fig output, savejpeg=0 to not save any .jpeg outputs and all_contribs_output=0 to not save the .txt file with a full breakdown of the spectrum. Leaving them all as 1 (except maybe the savejpeg) is strongly recommended.
Plot_Atomic_Contribs_V3
This is an optional script that plots certain breakdowns of your calculated DoS/XPS spectrum (i.e you run it AFTER Gen_Gelius_Spectrum) . Its not too hard to do the same thing in excel using the *ALL_Contribs* output file (see output file sections) but this is a lot faster. There are three types of breakdown this function can do (see pictures below):
1) Atomic breakdown with fragments kept seperate (merge_spd=1,merge_fragments=0)
2) Atomic orbital breakdown (merge_spd=0,merge_fragments=1)
3) Atomic breakdown (merge_spd=1,merge_fragments=1)
The function in its uploaded form takes a single argument , though there are two variables in the actual code that you need to mess with (merge_spd and merge_fragments).
So for example, to run this on the output file from the "Using the Example files" i typed into the matlab terminal:
Plot_Atomic_Contribs_V3('C:\Users\Richard\Desktop\Open_Shell_Code_Some_Stuff\Example_files\other_file\DoS_1ev_0pt3l')
The argument needs to be a folder containing the output files from the Gen_Gelius_Spectrum script. You'll then get a saved graph with the breakdown you asked for and also a .txt file containing the breakdown in a comma delimited format. The output file names record the options you used to make the script (e.g "Plotted_Atomic_Contribs_spd0_frag1.txt" means merge_spd was set to 0 and merge_fragments was set to 1).

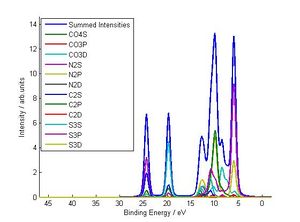
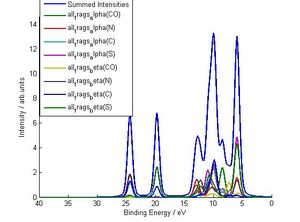
Output Files
The first "output file" is the *MCRF*.txt file, which just contains the commands to build the required fragment in multiwfn.
Files From Multiwfn
Most people should be able to completely ignore these files. But in case your curious:
1) *DOS_curve*.txt Essentially a DoS plot. 1st column is energies in au, second column is TDOS of the fragment. Not really sure what the 3rd column is. No particular use for this file, but it gets auto-generated by multiwfn and i dont go out my way to delete it.
2) *DOS_line*.txt Data to create a stick plot (where each MO is a stick). Plot column 1 vs column 2 as a line graph and you get an axis with sticks representing the MOs. 4th column onwards represent contribution of the various sub-fragments to each "stick". This is the file that gets used to generate the *ML_inpt.txt file.
3) *DOS_frag*.txt This file contains the sub-fragment info in a way multiwfn understands. If its renamed to DOS_frag.txt then multiwfn can import it to get the fragments (therefore can be useful for debugging, no further purpose).
MLinpt.txt File
Creation of the *ML_inpt.txt files is essentially the point of the gen_gelius_input* script. These are primarily input files for matlab, but they do have useful information in an easy to read format. If you import the file into excel and COMMA DELIMIT ONLY (i.e untick the space delimit option) you can get something like below(there are more decimal places in the actual file):
#M_FACTOR=1.0, #Energy(eV) TDOS CO4S CO3P CO3D N2S N2P N2D C2S C2P C2D S3S S3P S3D [LINES DELETED] -24.5 0.9 0.0 0.0 0.0 0.5 0.1 0.0 0.2 0.1 0.0 0.1 0.0 0.0 -24.4 1.0 0.0 0.0 0.0 0.5 0.1 0.0 0.3 0.1 0.0 0.0 0.0 0.0 -24.4 1.0 0.0 0.0 0.0 0.5 0.1 0.0 0.3 0.1 0.0 0.0 0.0 0.0 -24.4 1.0 0.0 0.0 0.0 0.5 0.1 0.0 0.3 0.1 0.0 0.0 0.0 0.0 -19.8 0.3 0.0 0.0 0.0 0.1 -0.1 0.0 0.2 0.1 0.0 0.7 0.1 0.0 -19.7 0.3 0.0 0.0 0.0 0.0 0.0 0.0 0.1 0.1 0.0 0.7 0.1 0.0 [LINES DELETED]
The shown file is "all_frags_alpha_MLinpt.txt", which is generated from the example files in the open shell code. As this actually comes from merging two MLinpt.txt files (gen_gelius_input.sh does this part silently) the TDOS in this case doesnt mean anything.
The first column lists the energy of each MO in eV (1 ev ~27.212 au). For any row, columns 3(CO4S)-->end (S3D) list the fractional contributions of each type of atmoic orbital to the MO in that row. For example the MO with energy -19.7 eV is mainly from S3s type atomic orbitals. These fractions were determiend by multiwfn by using Mulliken population analysis.
All the fractional contributions in the file get multiplied by the M_FACTOR value (as explained above in the Creating your own input files section). Therefore the value of M_FACTOR can easily be set after the MLinpt.txt file is created, as opposed to within the initial input file.
Formatted_UMOs
The default for the gen_gelius_input* script (i.e the settings for the version i uploaded) is to put only OCCUPIED MO energies/breakdowns into the *MLinpt.txt file. If you want all MOs (I really DONT recommend this) in the MLinpt.txt file then change all_orbs='cheese' to all_orbs='ALLORBS' at the top of the gen_gelius_input* script.
Otherwise you will get a *Formatted_UMOs* file for every MLinpt.txt file you get. The format of the file is identical to the MLinpt.txt format except the MO at the top of the file corresponds to the LUMO.
WARNING: If you use the link_files feature, then the output formatted_UMOs files DO NOT get merged. This shouldnt matter as you should never really use the formatted UMOs for anything.
Output From Matlab Code
The matlab code firstly will create a new folder with a name reflecting the input arguments. The format is hv_FWHM_%lorenztian where hv is the photon energy, FWHM is full width half maximum of the broadeing function and %lorenztian is the amount of lorenztian mixed in the broadening function. For example the earlier command (from using example files section) "Gen_Gelius_Spectrum(plist,-1,1)" generates the folder "DoS_1ev_0pt3l". DoS = Density of states, and means we did not use photoionisation cross sections. The 0pt3l part means our broadening function was 30% lorenztian and 70% Gaussian.
Two .fig files will have been produced from this code, these contain matlab graphs of the output. These .fig files follow the naming ("filename" is the name of the original fchk file):
1) filename_DoS.fig contains the total DoS plot.
2) filename_DoS_frag_contribs.fig contains the DoS AND the pDoS of each of the fragments. For closed shell code you get 1 fragment per input file (assuming you dont use link_files), but for the open shell code each input file leads to two fragments - one for alpha electrons and one for beta electrons.
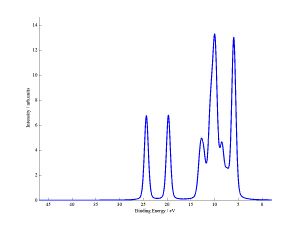
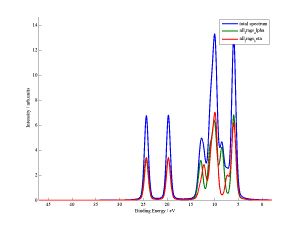
The filename_DoS.fig plot data is output in a .txt file of the same name, while each fragment in filename_DoS_frag_contribs.fig has its own .txt file. For the example above the two fragments have names "all_frags_alpha_other_file_DoS" and "all_frags_beta_other_file_DoS". The start of the names are taken from the name of the original input file "all_frags.test".
The final file output is the "ALL_Contribs_other_file_DoS.txt" file. This(as the name implies) contains all contributions to the DoS spectra in a comma delimited file. The Plot_Atomic_Contribs*.m code can be run on this file to generate plots with a few different breakdowns (e.g atomic breakdowns) - see the relevant section of this wiki (not done yet....). The file will look something like below (commas replaced with tabs):
#all_frags_alpha all_frags_beta #Sub fragments in each= 12 12 #Energy(eV) CO4S CO3P CO3D N2S N2P N2D C2S C2P C2D [COLUMNS REMOVED] [ROWS REMOVED,numbers rounded] 7.5 0.00 0.01 0.31 0.04 0.13 0.00 0.00 0.11 0.00 [COLUMNS REMOVED] 7.51 0.00 0.01 0.33 0.04 0.13 0.00 0.00 0.11 0.00 [COLUMNS REMOVED] 7.52 0.00 0.01 0.34 0.04 0.13 0.00 0.00 0.11 0.00 [COLUMNS REMOVED] 7.53 0.00 0.01 0.35 0.04 0.14 0.00 0.00 0.12 0.00 [COLUMNS REMOVED] 7.54 0.00 0.01 0.36 0.04 0.14 0.00 0.00 0.12 0.00 [COLUMNS REMOVED] 7.55 0.00 0.02 0.37 0.04 0.14 0.00 0.00 0.12 0.00 [COLUMNS REMOVED] 7.56 0.00 0.02 0.39 0.05 0.14 0.00 0.00 0.13 0.00 [COLUMNS REMOVED] [ROWS REMOVED]
Hopefully the format is mostly intuitive, the top two rows contain information on what the data is, and the rest contain the data. The top row contains (originally comma delimited) a list of all "fragments" names, which is the number of input files for the closed shell code and 2x the number of input files for the open shell code. The second row contains the number of sub-fragments (which should be atomic orbitals) in each fragment. In the example above each fragment had 12 types of atomic orbital.
So for the example above column 2-14 shows contributions from "all_frags_alpha" while column 15-27 shows contributions from "all_frags_beta" (column 1 lists energies).
Bugs/Things to Watch For
Fragment Names
Make sure your fragment names arent a substring of your .fchk file names. For example an input file called "Cl.test" wont work for a file called "BMIM_Cl.fchk". However, "Cl_frag.test" will work. Therefore personally i add _frag to the end of every input file name. The other way to sort this is to set linked_files=0 in gen_gelius_input_opensh.sh, sincce the link files part of the code is where this bug comes from.
Fragment Error Checking
Unless you manually turn it off (The "error=" option in gen_gelius_input_opensh.sh script) the code tries to check your input file for errors. Specifically it tests whether the TDOS in an input_file really is the sum of the other sub-fragments in the input(.test) file. E.g it will trigger if you request only Sulfur p-type and d-type AOs (but no s-type AOs) in the input file, or if you request all sulfur p-type AOs twice. If the error check triggers then it BREAKS the input file for matlab by putting an error msg in the first line of the *MLinpt.txt file. An example of what the error looks like is:
“Fragment Sum incorrect on row 19 for fchk file xxxx.fchk”
If you *MLinpt.txt files arent working in matlab then this is probably the reason.
Pathlengths
This code makes a lot of sub-folders based on the name of .fchk files. If you have really long .fchk file names its likely that you'll end up with some files which exceed the 256 character limit for a pathlength on a windows computer. In matlab 2014 this will cause the code to exit with an error, saying it cant save the .fig files. In newer matlab versions it may just save the output files , but you wont be able to open them on windows if the path length is greater than 256 characters. Ways to avoid this are to shorten .fchk file names where possible, or (less practical) run the code on a folder as close as possible to root (i.e a folder with a short a pathlength as possible).
