Talk:Mh3413CMP
JC: General comments: Report is well written with clear graphs of results. Code is also clear and well commented. Make sure that figures appear at the most appropiate point of the text and a few explanations could have been expanded on a bit.
Monte Carlo Simulations of the Ising Model
Abstract
By applying a Monte Carlo algorithm to a square lattice of magnetic atoms in the Ising model, the properties of a two-dimensional ferromagnetic system were investigated. It was found that below the Curie temperature the whole lattice would spontaneously align to produce a net magnetization in the presence of no external magnetic field. At a reduced temperature of T≈2.0 for the majority of lattice sizes, a sharp decrease in the magnetisation, along with an increase in the volatility of magnetization and energy was observed, corresponding to the Curie temperature. As the temperature of the system increased for all lattice sizes, the energy per spin plateaued to a maximum value and the magnetization equilibrated to a value of zero, resembling a function. Using curve polynomial fitting techniques for the experimental data, the for all the lattice sizes were able to be established and used to establish the value of . The Curie temperature was observed to decrease with increasing lattice size.
Overview of Ising Model
The Ising model is a simple physical model describing the ferromagnetic properties of a regular square lattice system, in the presence of no external magnetic field. In this model, the vectors of the atomic magnetic dipoles are restricted to states of pointing up(+1) or down(-1) in the two dimensional structure. The equation in Figure 1 describes the interaction energy in a system upon the assumption of:
- Only short range, nearest neighbor interactions between the adjacent lattice cells contributing to the total energy
- Periodic boundary conditions applied to the lattice structures, causing them to wrap on them themselves
- No external magnetic field applied
| Figure 1: The energy of the interactions with the adjacent spins of lattice cell, when no magnetic field is applied |
The J is a coupling constant and is a quantitative description of the level of interaction between the adjacent spin cells. It was assumed that J=1.0 in the follow sections, resulting in the energy being in reduced units of .
 |
|---|
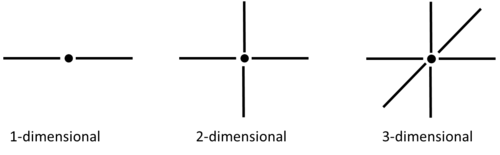
| Figure 2: Number of nearest neighbours for each lattice dimension |
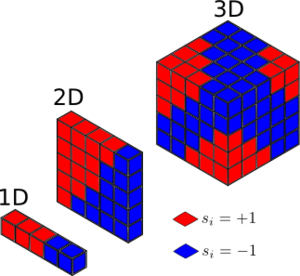
TASK: For the one dimensional case, there are two nearest neighbours with respect to the central atom. For 2- and 3-dimensional lattice systems, there are 4 and 6 respective neighbors. Overall, in a lattice with periodic boundary conditions, each lattice cell would have , nearest neighbours (where d=the number of dimensions). In the following model, it was assumed that the lattice was a 2-dimensional system.
Using the equation in figure 1, the lowest energy state would correspond to a system where all the spins of the atoms are in the same state (either up or down): The factor of 1/2 is to account for the double counting of the interactions between the lattice atoms.
JC: Correct, try to be consistent using either d or D for dimensions.
TASK: As all the atoms have the same spin state, the multiplicity of the system is 1, there is only one way to arrange the system. This system would be present at a temperature of 0K.
This result is consistent with the 3rd law of thermodynamics which states that "the entropy of a perfectly crystalline system is 0 at a temperature of 0K".
JC: Why do you not count the all spin up and all spin down states separately?
TASK: The change in spin from -1 to +1 for one of the cells can be thought of as two processes: Change from -1 to 0 and 0 to +1 spin For a () system in the lowest energy configuration, when one of the spins spontaneously change direction the energy change would correspond to:
The entropy of this system would increase, as there would be an increase in the number of ways to produce the state (increase in multiplicity).
JC: Correct, good reasoning.
The magnetisation of the system is calculated by the sum of the parallel spins in the system. In the lowest energy configuration, where all the atoms in the lattice have the same direction of spin, the system would expect to have a maximum value of magnetisation. This corresponds to high ferromagnetic state[1].
TASK: For figure 3, the magnetisation for the 1D and 2D case would be expected to be , as the number of up spin states are greater than the number of down spin states. As there is no external magnetic field that is present in the system, the sign of magnetization is not significant.
TASK: For a () system where the atomic magnetic dipoles do not have sufficient energy to repel against their neighbours, it would be expected that the cells would be all aligned in the same direction. This would be most favourable in energetic terms and lead to a maximum magnetisation value of
JC: Correct, but remember could be + or -.
Calculating the energy and magnetisation
TASK: It was assumed that J=1.0 in the follow sections. When J>0 there is a energetic drive for the spins to align as this minimizes the energy of the system.[2] The figure 4 below, presents the code that initally was used to calculate the energy and the magnetisation of the system. Please refer to the Accelerating Code section for the modified version of the script.
Energy
The figure 5 gives a visual illustration of the actions undertaken on the randomly generated original matrix. These actions were undertaken to ensure the final energy calculations of the matrix took into consideration the assumption of the periodic boundary conditions, and for the simplification of the energy calculations.
The 1st matrix that is printed in figure 5 is the original matrix random generated from [-1,1] states. The energy function takes copies of the 1st and last rows of the original matrix and creates a new matrix, where the copied last row of the original matrix, becomes the 1st row of the new matrix. Likewise, the 1st row of the original matrix becomes the last row of the new matrix. The original x matrix within this additional 'wall' of numbers was left untouched. This same operation was applied to the columns, such that a 6x6 matrix was formed. The calculation of the interaction energies was only applied to the original 4x4 matrix within the 'wall' of the copied values, using the equation in figure 1.
Magnetisation
As the matrix values represent the magnetic spin of atoms in a lattice, the values are restricted to [-1,1]. The sum of the spins values in the lattice is equal to the total magnetisation of the system; a simple sum(sum(x)) was applied to the matrix to return the total summation value. The model assumes that in the presence of no external magnetic field, the lattice can still generate a net magnetisation through the long range correlations.
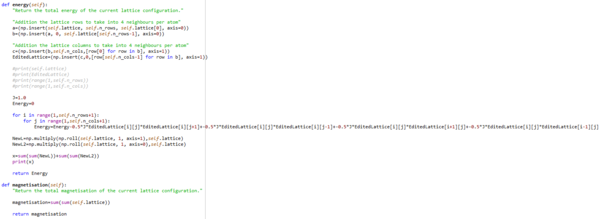
TASK: The main engine of the model that initally was used to calculate the energy and magnetisation. Part of the IsingLatticeModel script.
def energy(self):
"Return the total energy of the current lattice configuration."
"Addition the lattice rows to take into 4 neighbours per atom"
a=(np.insert(self.lattice, self.n_rows, self.lattice[0], axis=0))
b=(np.insert(a, 0, self.lattice[self.n_rows-1], axis=0))
"Addition the lattice columns to take into 4 neighbours per atom"
c=(np.insert(b,self.n_cols,[row[0] for row in b], axis=1))
EditedLattice=(np.insert(c,0,[row[self.n_cols-1] for row in b], axis=1))
#print(self.lattice)
#print(EditedLattice)
#print(range(1,self.n_rows))
#print(range(1,self.n_cols))
J=1.0
Energy=0
for i in range(1,self.n_rows+1):
for j in range(1,self.n_cols+1):
Energy=Energy-0.5*J*EditedLattice[i][j]*EditedLattice[i][j+1]+-0.5*J*EditedLattice[i][j]*EditedLattice[i][j-1]+-0.5*J*EditedLattice[i][j]*EditedLattice[i+1][j]+-0.5*J*EditedLattice[i][j]*EditedLattice[i-1][j]
return Energy
def magnetisation(self):
"Return the total magnetisation of the current lattice configuration."
magnetisation=sum(sum(self.lattice))
return magnetisation
|
JC: Better to write the spin values as {1, -1}, putting them in square brackets suggests that the spin can take any value in the interval 1 to -1. Energy code looks good, nice method to take account of periodic boundary conditions.
Introduction to Monte Carlo simulation
TASK: Please refer to the Accelerated Code section to see the modified IsingLattice code
TASK: Due to the spin up(+1) and spin down(-1) possibilities for each atom, there are a increasing number of configurations possible with the increasing number of atoms.
For a 100 spin system and computing power of calculations per second:
JC: Correct, can you write this in more intuitive units, e.g. years/centuries? Code looks good and is well commented.
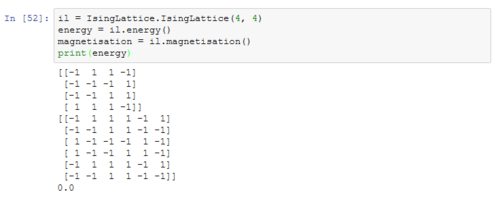
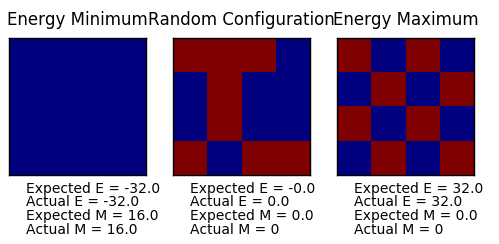
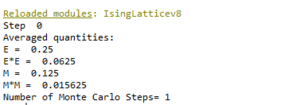
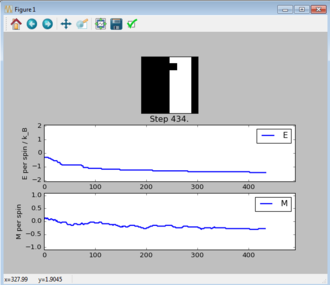
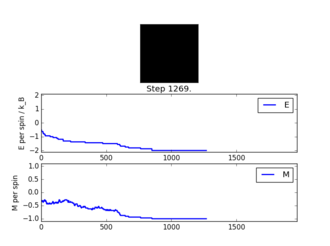
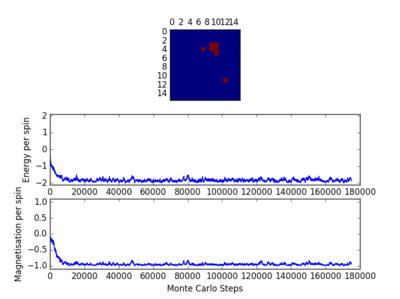
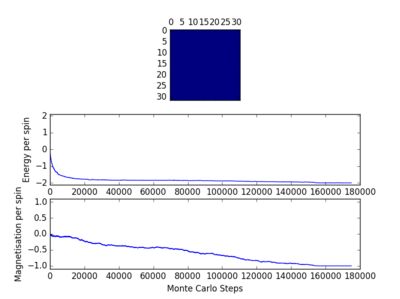
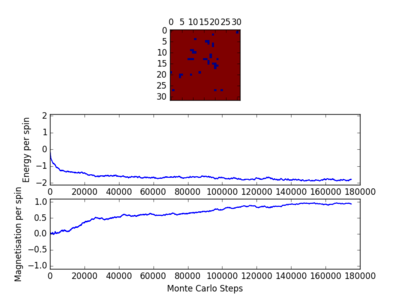
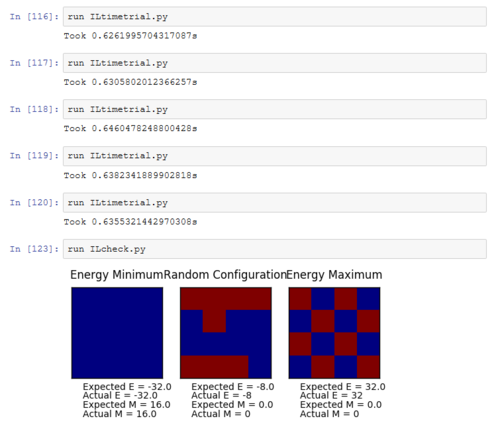
TASK: Figure 7 to 10 captures the development and testing of the montecarlocycle() and statistics() function. Figures 9 and 10 ultilised the ILanim.py script to animate a real time Monte Carlo simulation. Figure 9 presents the minimum energy and maximum magnetisation value of the system in equilibrium state (ie graphs of the energy and magnetisation per spin). It is expected that when , there would be spontaneous magnetisation (i.e. ) as the exchange and long range correlation effects would outweigh the thermal fluctuations, causing the neighboring atoms to favour aligning in the same direction. Above the , the thermal energy in the system is sufficient to overcome any exchange and long range correlation effects.
Figure 10 is a system in an equilibrium state.
Accelerating the code
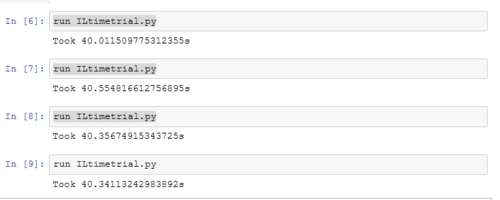
TASK: Using the script ILtimetrial.py, the time required to perform 2000 Monte Carlo steps in the IsingLattice.py script was calculated. On average the original code took:
The code was optimized using the roll() and multiply() functions. The original matrix was multiplied with the original matrix shifted by one unit in the x-axis and the original matrix shifted by one unit in the y-axis. This operation was able to capture the same neighbouring interactions as the previous code, but was more time efficient. On average the optimized code took:
JC: How many runs were used to calculate the average time taken?
 |
 |
|---|---|
| Figure 11: Before Optimization of energy and magnetization code | Figure 12: After Optimization of energy and magnetization code |
Optimized Ising Lattice Code
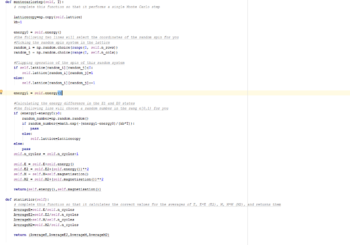
TASK: The main engine of the model that applies the Monte Carlo steps and calculates the energy and magnetisation of the system
import numpy as np
import math as math
class IsingLattice:
E = 0.0
E2 = 0.0
M = 0.0
M2 = 0.0
NoIgnore = 160000
n_cycles = 0
def __init__(self, n_rows, n_cols):
self.n_rows = n_rows
self.n_cols = n_cols
self.lattice = np.random.choice([-1,1], size=(n_rows, n_cols))
def energy(self):
"Return the total energy of the current lattice configuration."
J=1.0
Energy=0
NewL=np.multiply(np.roll(self.lattice, 1, axis=1),self.lattice)
NewL2=np.multiply(np.roll(self.lattice, 1, axis=0),self.lattice)
Energy=(sum(sum(NewL))+sum(sum(NewL2)))*-J
return Energy
def magnetisation(self):
"Return the total magnetisation of the current lattice configuration."
magnetisation=sum(sum(self.lattice))
return magnetisation
def montecarlostep(self, T):
# complete this function so that it performs a single Monte Carlo step
latticecopy=np.copy(self.lattice)
kb=1
energy0 = self.energy()
#the following two lines will select the coordinates of the random spin for you
#Picking the random spin system in the lattice
random_i = np.random.choice(range(0, self.n_rows))
random_j = np.random.choice(range(0, self.n_cols))
#Flipping operation of the spin of this random system
if self.lattice[random_i][random_j]<0:
self.lattice[random_i][random_j]=1
else:
self.lattice[random_i][random_j]=-1
energy1 = self.energy()
#Calculating the energy difference in the E1 and E0 states
#the following line will choose a random number in the rang e[0,1) for you
if (energy1-energy0)>0:
random_number=np.random.random()
if random_number<=math.exp(-(energy1-energy0)/(kb*T)):
pass
else:
self.lattice=latticecopy
else:
pass
self.n_cycles = self.n_cycles+1
if self.n_cycles>=self.NoIgnore:
self.E = self.E+self.energy()
self.E2 = self.E2+(self.energy())**2
self.M = self.M+self.magnetisation()
self.M2 = self.M2+(self.magnetisation())**2
else:
self.E = 0
self.E2 = 0
self.M = 0
self.M2 = 0
return(self.energy(),self.magnetisation())
def statistics(self):
# complete this function so that it calculates the correct values for the averages of E, E*E (E2), M, M*M (M2), and returns them
if self.n_cycles>=self.NoIgnore:
AverageE=self.E/(self.n_cycles-self.NoIgnore)
AverageE2=self.E2/(self.n_cycles-self.NoIgnore)
AverageM=self.M/(self.n_cycles-self.NoIgnore)
AverageM2=self.M2/(self.n_cycles-self.NoIgnore)
else: #if not above the number of cycles ignored, the system will ignore the value
AverageE=0
AverageE2=0
AverageM=0
AverageM2=0
# take into consideration the ignored cycles in the printing of the number of steps undertaken
return (AverageE,AverageE2,AverageM,AverageM2,(self.n_cycles-self.NoIgnore))
|
The effect of temperature
As the model starts from a random set of lattice spins, there is a certain number of steps required in order for the system to reach an energetic and magnetic thermal equilibrium. There are two main variables that effect this value:
- Lattice Size
- Temperature
Note: In each of the captions of the figures below, there is an approximated number of steps required to reach the equilibrium state
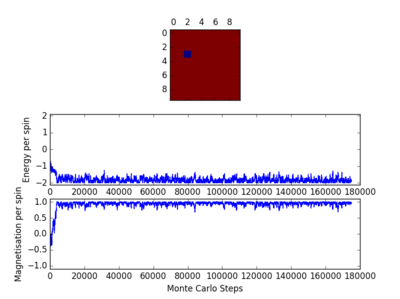
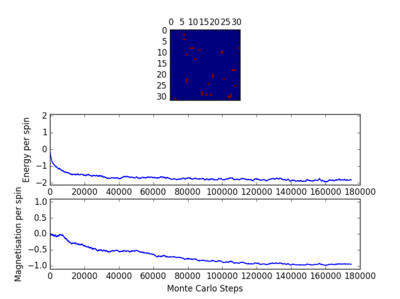
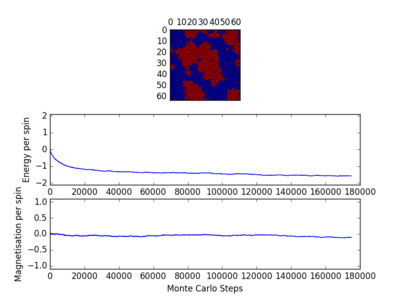
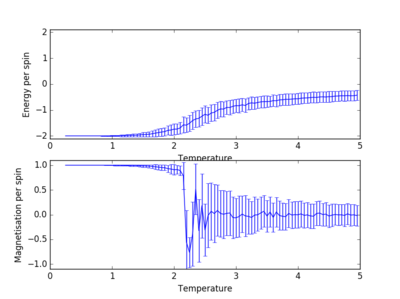
For figures 13 to 16 the temperature was kept constant at T=1.85, while the lattice size was increased. As seen below, with the increasing lattice sizes, the number of steps required to reach the equilibrium position increased. For example, the magnetisation and energy in the 10x10 lattice structure equilibrised <10000 steps, while in the 64x64 system the energy reached equilibrium at approx 160,000 steps.
Changing Lattice Size
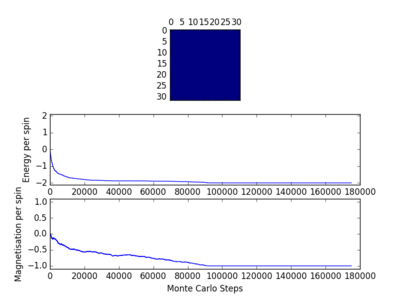
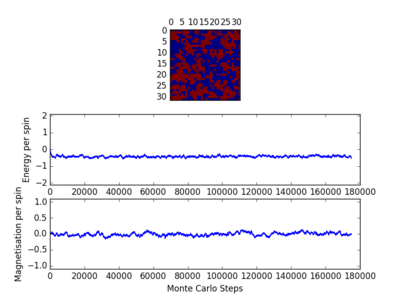
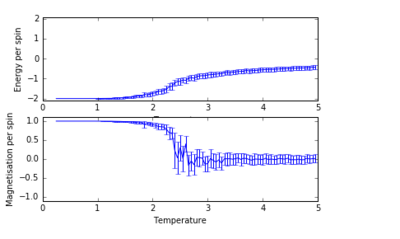
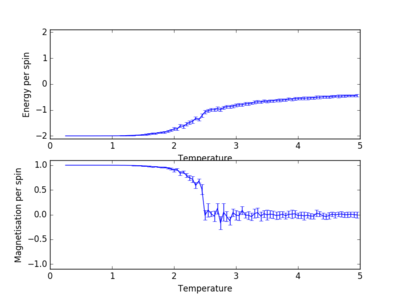
For figures 17 to 20, the lattice size was kept constant at 32x32, while the reduced temperatures were varied between T=0.25-5.0. With increasing temperatures of the system, the number of steps required to reach the equilibrium position in the system increased. For figure 20, T=5 is significantly greater than the Curie temperature for the system and thus the system reaches magnetic and energy equilibrium instantaneously as the thermal fluctuations eliminate all the correlation, enthaplic and entropic factors.
Changing Temperature
TASK: It was found that for each lattice size and temperature, the system has a different number of cycles needed to obtain an equilibrium state. Based on the information above, for the worst case scenarios (T= approx 1.85 and very large lattice sizes eg 64x64), all the lattices reach an equilibrium by 160,000 steps. Therefore, the first 160,000 cycles of the Monte Carlo steps were chosen to be ignored in the calculation of the averages. This result led to the average values of magnetisation and energy having greater magnitudes and more accurate values for approximating the equilibrium system. A greater number of steps could have been ignored, and this may have lead to increased accuracy in the calculation of the variables, however in terms of the computational time required to calculate the these variables (eg for runtime=240,000) and the benefit obtained, it was decided that an optimal value of steps to ignore was 160,000.
The Ising Lattice script model no longer considers the energy and magnetisation values for the steps up to 160,000 ie until the system reaches an equilibrium point.
JC: Good thorough choice of number of steps to ignore and correctly introduced into code.
Running over a range of temperatures/Scaling the System Size
TASK: Using the ILtemperaturerange.py script, the average energy and magnetisation was plotted against the reduced temperature. Taking into account the 160,000 ignored simulations, the runtime was set to 240,000 and the dt steps adjusted to a lower value of 0.05 over a temperature range from T=0.25-5.0. This allowed the fine details of the fluctuations in energy and magnetisation for the system to be captured. The error bars at each temperature point, are calculated using the fluctuations in values from the runs conducted. The variability originates from the variations that occur in the .
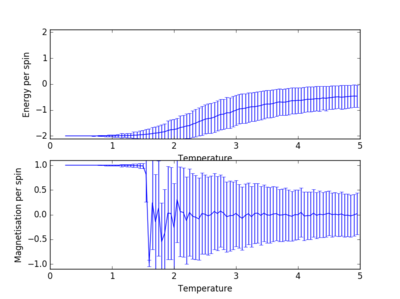
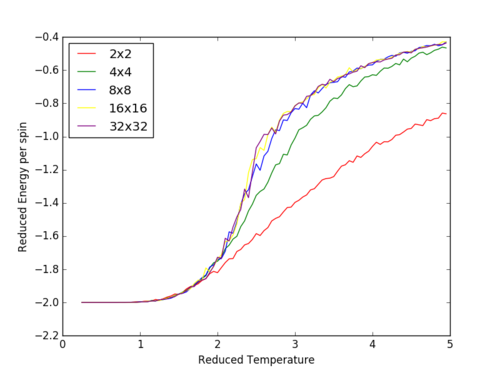
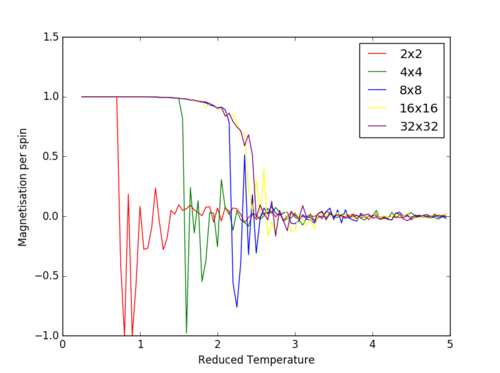
The Curie Temperature is the temperature at which the system undergoes a phase transition from an ordered to a disordered system and loses the net magnetization. The magnetisation changes rapidly at this point. Above the Curie Temperature, the changing of the spins from an ordered system to a completely disordered system – there is no energy penalty involved in this. In addition, there are long range correlation interactions present in the system which result a spin change, increasing the probability of triggering a spin change in another atom. Figure 25 captures the energy per spin against the reduced temperature. The energy rapdily increases upon reaching the Curie temperature; this graph resembles a function. With increasing lattice size, the energy per spin was observed to increase, with the 8x8,16x16 and 32x32 having very similar shapes.
The Curie temperature was observed to increase with increasing lattice sizes in figure 26, but contradicted the results of figure 25 in which the point of inflexion (steepest gradient for dE/dT) corresponds to the Curie temperature; the Curie temperature decrease with increasing lattice size. It is believed that for figure 26, the small size of the lattice caused inaccuracies in the simulations, due to the each lattice cell being so close to one another. The change in a spin of one cell in this lattice would dramatically increase the probability of the neighboring lattice cells changing spins. Furthermore, if the lattice cell spends half of its time in a total spin up state and the other half of its time in a total spin down state, the average of the system would end up being around zero.
JC: Graphs look good. How does the energy against temperature graph resemble a curve? Is there a theoretical justification for this? In the small lattice, flipping a single spin is more likely to cause the entire lattice to flip.
TASK: Furthermore, the increased volatility around the Curie Temperature point decreases in significance with increasing lattice sizes (Figure 26). The temperature range at which the phase transition takes place, decreases with increasing lattice size. For a 32x32 system, the phase transition takes place almost instantly with insignificant volatility, while in the 4x4 system the transition takes place over approximately T=3 due to the fluctuation within the system. It is believed that for a lattice size of 16x16, the system is large enough to capture the long range fluctuations due to the sharp transition and the fluctuations observed.
 |
|
|---|---|
| Figure 25: Reduced energy per spin versus reduced temperature | Figure 26: Reduced magnetisation per spin versus reduced temperature |
JC: The energy per spin curves seem to have converged for the 8x8 system..
Running over a range of temperatures
TASK: Plotting of the average energy and magnetisation for each temperature, with error bars, for an 8x8 lattice
from IsingLattice import *
from matplotlib import pylab as pl
import numpy as np
n_rows = 8
n_cols = 8
il = IsingLattice(n_rows, n_cols)
il.lattice = np.ones((n_rows, n_cols))
spins = n_rows*n_cols
runtime = 240000
times = range(runtime)
temps = np.arange(0.25, 5.0, 0.05)
#Empty arrays to be filled
energies = []
magnetisations = []
energysq = []
magnetisationsq = []
stdenergy = []
stdmagnetisation=[]
for t in temps:
for i in times:
if i % 100 == 0:
print(t, i)
energy, magnetisation = il.montecarlostep(t)
aveE, aveE2, aveM, aveM2, n_cycles = il.statistics()
energies.append(aveE)
energysq.append(aveE2)
magnetisations.append(aveM)
magnetisationsq.append(aveM2)
#reset the IL object for the next cycle
il.E = 0.0
il.E2 = 0.0
il.M = 0.0
il.M2 = 0.0
il.n_cycles = 0
#Calculation in the energy and magnetisation errors of the system by calculating the Var(x) and then the standard deviations
energyerror=np.array(energysq)-(np.array(energies)**2)
magnetisationserror=np.array(magnetisationsq)-(np.array(magnetisations)**2)
stdmagnetisation=np.sqrt(magnetisationserror)
stdenergy=np.sqrt(energyerror)
#Ploting figure and formatting issues
fig = pl.figure()
enerax = fig.add_subplot(2,1,1)
enerax.set_ylabel("Energy per spin")
enerax.set_xlabel("Temperature")
enerax.set_ylim([-2.1, 2.1])
magax = fig.add_subplot(2,1,2)
magax.set_ylabel("Magnetisation per spin")
magax.set_xlabel("Temperature")
magax.set_ylim([-1.1, 1.1])
#Addition of error bars for each of the temperature points run at
enerax.errorbar(temps, np.array(energies)/spins,yerr=np.array(stdenergy)/spins)
magax.errorbar(temps, np.array(magnetisations)/spins,yerr=np.array(stdmagnetisation)/spins)
pl.show()
final_data = np.column_stack((temps, energies, energysq, magnetisations, magnetisationsq,stdmagnetisation,stdenergy))
np.savetxt("32x32-larger dt.dat", final_data)
|
Scaling the System Size
TASK: The code below was used to plot energy and magnetisation per spin against temperature. Currently, it plots the energy but can be easily adapted for magnestism.
import matplotlib.pyplot as plt
import numpy as np
#Extracting the data from each data file and catergorising it into variables
data2by2 = np.loadtxt("2x2.dat")
spins2by2=4
temps2by2=data2by2[:,0]
energies2by2=data2by2[:,1]
energysq2by2=data2by2[:,2]
magnetisations2by2=data2by2[:,3]
magnetisationsq2by2=data2by2[:,4]
data4by4 = np.loadtxt("4x4.dat")
spins4by4=16
temps4by4=data4by4[:,0]
energies4by4=data4by4[:,1]
energysq4by4=data4by4[:,2]
magnetisations4by4=data4by4[:,3]
magnetisationsq4by4=data4by4[:,4]
data8by8 = np.loadtxt("8x8.dat")
spins8by8=64
temps8by8=data8by8[:,0]
energies8by8=data8by8[:,1]
energysq8by8=data8by8[:,2]
magnetisations8by8=data8by8[:,3]
magnetisationsq8by8=data8by8[:,4]
data16by16 = np.loadtxt("16x16.dat")
spins16by16=256
temps16by16=data16by16[:,0]
energies16by16=data16by16[:,1]
energysq16by16=data16by16[:,2]
magnetisations16by16=data16by16[:,3]
magnetisationsq16by16=data16by16[:,4]
data32by32 = np.loadtxt("32x32.dat")
temps32by32=data32by32[:,0]
spins32by32=1024
energies32by32=data32by32[:,1]
energysq32by32=data32by32[:,2]
magnetisations32by32=data32by32[:,3]
magnetisationsq32by32=data32by32[:,4]
#graph curves colour selection
plt.gca().set_color_cycle(['red', 'green', 'blue', 'yellow', 'purple'])
#Calculation of the properties per spin and plotted
plt.plot(temps2by2, energies2by2/spins2by2)
plt.plot(temps4by4, energies4by4/spins4by4)
plt.plot(temps8by8, energies8by8/spins8by8)
plt.plot(temps16by16, energies16by16/spins16by16)
plt.plot(temps32by32, energies32by32/spins32by32)
plt.xlabel('Reduced Temperature')
plt.ylabel('Reduced Energy per spin')
plt.legend(['2x2', '4x4', '8x8', '16x16','32x32'], loc='upper right')
plt.show()
plt.savefig('test.jpg')
|
JC: Code looks good, good use of the loadtxt function.
Determining the heat capacity
TASK: Proof of:
=
LDS:
Or
where
JC: Shouldn't have second sum in this equation.
Using the quotient rule:
JC: Careful with the brackets, the sum should be inside the brackets for the last term.
RDS:
Calculating the heat capacity
=
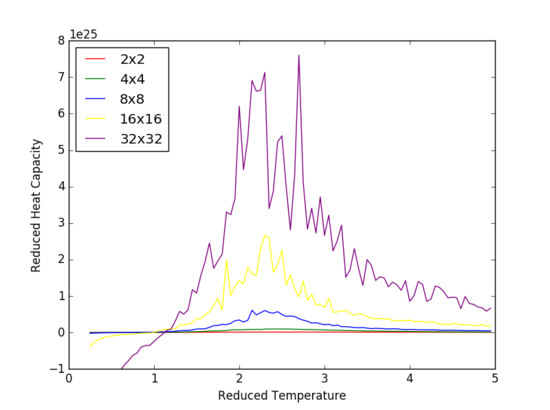
TASK: The increased detail and volatility around the curve at the Curie Temperature, is due to the lower value of dt=0.05 that was run in the 'Running over a range of temperatures' section. This section utilized the .dat files that were produced from the previous tasks. The heat capacity vs reduced temperature graph presents a strong peak in vicinity of the critical temperature, caused by the , the gradient of the energy per spin vs reduced temperature rapidly increasing near the Curie temperature. The point of inflexion in this graph (energy per spin vs reduced temperature) corresponds to the maxima peak observed in the heat capacity versus temperature graph. It is shown that with increasing array size, an increase in the peak amplitude is observed.
Calculating the heat capacity
TASK: Plot showing the heat capacity versus temperature for each of the lattice sizes from the previous section
import matplotlib.pyplot as plt
import numpy as np
#Load the data from the temperature range files and calculate the variance of each of the energies using the quoted formula
data2by2 = np.loadtxt("2x2.dat")
spins2by2=4
temps2by2=data2by2[:,0]
Varenergies2by2=data2by2[:,2]-data2by2[:,1]**2
data4by4 = np.loadtxt("4x4.dat")
spins4by4=16
temps4by4=data4by4[:,0]
Varenergies4by4=data4by4[:,2]-data4by4[:,1]**2
data8by8 = np.loadtxt("8x8.dat")
spins8by8=64
temps8by8=data8by8[:,0]
Varenergies8by8=data8by8[:,2]-data8by8[:,1]**2
data16by16 = np.loadtxt("16x16.dat")
spins16by16=256
temps16by16=data16by16[:,0]
Varenergies16by16=data16by16[:,2]-data16by16[:,1]**2
data32by32 = np.loadtxt("32x32.dat")
temps32by32=data32by32[:,0]
spins32by32=1024
Varenergies32by32=data32by32[:,2]-data32by32[:,1]**2
plt.gca().set_color_cycle(['red', 'green', 'blue', 'yellow', 'purple'])
#plotting of the Variance/kT2 against the reduced temperature
plt.plot(temps2by2,Varenergies2by2/np.multiply(1.38064852e-23,temps2by2**2))
plt.plot(temps4by4, Varenergies4by4/np.multiply(1.38064852e-23,temps2by2**2))
plt.plot(temps8by8,Varenergies8by8/np.multiply(1.38064852e-23,temps2by2**2))
plt.plot(temps16by16, Varenergies16by16/np.multiply(1.38064852e-23,temps2by2**2))
plt.plot(temps32by32, Varenergies32by32/np.multiply(1.38064852e-23,temps2by2**2))
plt.xlim((0,5))
plt.ylim((-1E+25,0.8E+26))
plt.xlabel('Reduced Temperature')
plt.ylabel('Reduced Heat Capacity')
plt.legend(['2x2', '4x4', '8x8', '16x16','32x32'], loc='upper left')
plt.show()
|
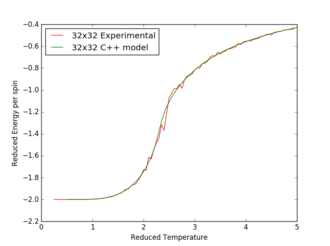
Locating the Curie temperature
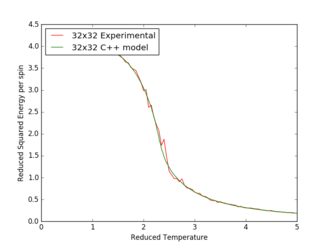
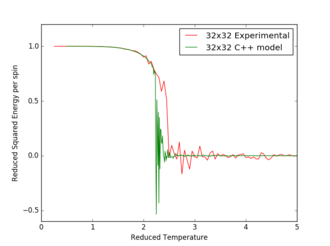
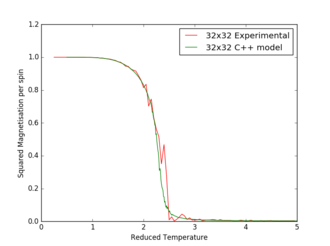
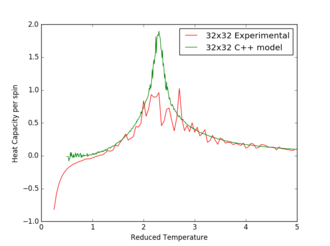
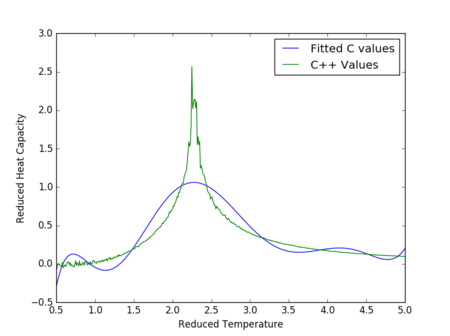
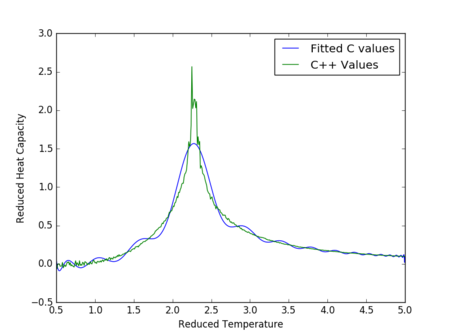
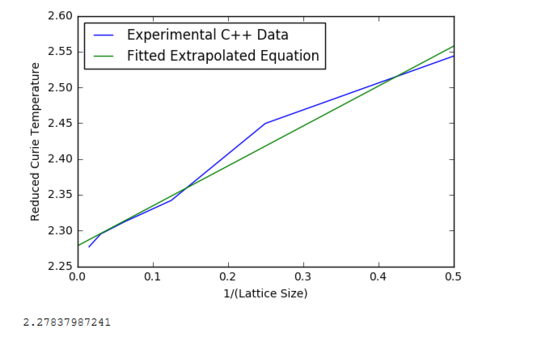
Figures 17 to 21 are the plots of the 32x32 lattice C++ data with the computational experimental data obtained for the per spin variables. The 32x32 lattice was used for the data comparison, as the volatility and error associated with the energy and magnetisation calculations for this lattice size was significantly lower than for the smaller arrays. A legend was added to the graphs to indicate the source of data. Overall, there is a good agreement between the C++ and the experimental data, however the C++ calculation method does show advantages in figure 31. The C++ data produces a pronounced peak, indicating a clear , while the experimental data has large variability around this temperature point. This accuracy can be attributed to the large number of steps that the C++ data undertakes.
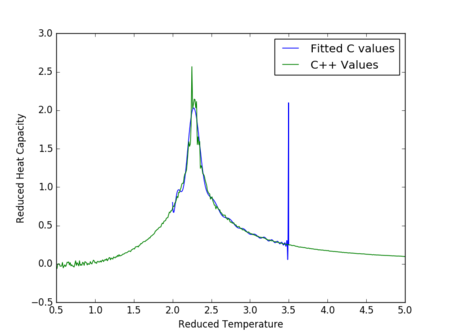
The figures 32-34 present the 'trial and error' method was applied to a 64x64 lattice, to produce the polynominal fit to the C++ data. It was found that a maximum polynominal value of 400 produced the best fit for the whole temperature range. However, due to the nature of the peak being very abrupt in the function, the polynominal fit function was unable to produce a satisfactory fit. When the temperature range of the polynominal fit function was reduced to the regions of the peak, the polynominal was able to produce a good fit (figure 34).
TASK:
Comparing the C++ data and the experimental data
TASK: For each lattice size, plotting the C++ data against experimental data. This current code is optimized for comparing the heat capacity values across the temperature range, but can be made to plot for
import matplotlib.pyplot as plt
import numpy as np
#My experimental data, calculation of the variance of the energy to use in the equation for the heat capacity
data32by32 = np.loadtxt("32x32.dat")
spins32by32=32*32
temps32by32=data32by32[:,0]
Varenergies32by32=data32by32[:,2]-data32by32[:,1]**2
#C++ data provided
Cdata32by32 = np.loadtxt("32x32c++.dat")
Cspins32by32=32*32
Ctemps32by32=Cdata32by32[:,0]
Cenergies32by32=Cdata32by32[:,5]
#print(Cenergies32by32)
plt.gca().set_color_cycle(['red', 'green', 'blue', 'yellow', 'pink'])
#calculation of the actual heat capacity ie mulitplied by kb + plotting the data on the same graph
plt.plot(temps32by32, (1.38064852e-23*Varenergies32by32/np.multiply(1.38064852e-23,temps32by32**2))/spins32by32)
plt.plot(Ctemps32by32,Cenergies32by32)
plt.xlabel('Reduced Temperature')
plt.ylabel('Heat Capacity per spin')
plt.legend(['32x32 Experimental', '32x32 C++ model'], loc='upper right')
plt.show()
|
Comparing C++ data and experimental data + polynominal fitting
TASK: A script to read the data from a particular file, and plot C vs T, as well as a fitted polynomial. The 400 degree polynomial was found to be the best fit without restricting the temperature ranges. Figures 32-33
TASK: Modified version of the previous script to fit the polynomial to the peak of the heat capacity ie restricting the temperature range figures 34
import matplotlib.pyplot as plt
import numpy as np
#List for the values of the maximum heat capacity and temperature to go into
Temperature=[]
HeatCapacity=[]
length=[2,4,8,16,32,64]
name=["2x2c++","4x4c++","8x8c++","16x16c++","32x32c++","64x64c++"]
#Loop to export data intp python from various c++ data files
for i in name:
naming= i+".dat"
data = np.loadtxt(str(naming)) #assume data is now a 2D array containing two columns, T and C
T = data[:,0] #get the first column
C = data[:,5] # get the 6th column
#The region which the polynominal will fit to
Tmin = 1.5
Tmax = 4.0
#Restricting the peak picking to the temeprature peak region due to peaks at the edge of boundaries
#Restricting Tmin and Tmax to other values if produces a better fit to the data
if i=="2x2c++":
Tminforpeak = 1.5
Tmaxforpeak = 3.0
elif i=="4x4c++":
Tminforpeak = 1.7
Tmaxforpeak = 3.3
elif i=="8x8c++":
Tminforpeak = 1.8
Tmaxforpeak = 3.3
elif i=="16x16c++":
Tmin = 1.5
Tmax = 2.9
Tminforpeak = 1.5
Tmaxforpeak = 2.8
elif i=="32x32c++":
Tmin = 1.5
Tmax = 2.8
Tminforpeak = 1.5
Tmaxforpeak = 2.7
else:
Tmin = 1.5
Tmax = 2.75
Tminforpeak = 1.5
Tmaxforpeak = 2.7
selection = np.logical_and(T >=Tmin, T <= Tmax) #choose only those rows where both conditions are true
peak_T_values = T[selection]
peak_C_values = C[selection]
#first we fit the polynomial to the data
fit = np.polyfit(peak_T_values, peak_C_values, 400) # fit a third order polynomial
#now we generate interpolated values of the fitted polynomial over the range of our function
T_range = np.linspace(Tminforpeak , Tmaxforpeak, 1000) #generate 1000 evenly spaced points between T_min and T_max
fitted_C_values = np.polyval(fit, T_range) # use the fit object to generate the corresponding values of C
#print(T)
Cmax = np.max(fitted_C_values)
Tmax = T_range[fitted_C_values == Cmax]
#Exporting the Heat Capacity maximum, temperature value and the corresponding lattice size to a .dat file
Temperature.append(Tmax)
HeatCapacity.append(Cmax)
final_data = np.column_stack((np.array(length),np.array(Temperature), np.array(HeatCapacity)))
np.savetxt("Heat Capacity Ising Model.dat", final_data)
|
Finding the peak in C
TASK:
Using the equation above, the 1/(lattice length) was plotted against the reduced temperature. The resulting linear relationship was then extrapolated and calculated at T=0; corresponds to the value of . This was compared with Onsager(1944[3]) literature values and had very good agreement.
The major sources of error are due to the assumptions made in the model:
- The noise at the peak value creates uncertainty in the plot fitting
- Assumption that the relationship in figure 35 is linear
- Required to extrapolation data for T=0 - uncertainty in what the actual data would be in this region
- There are more than 4 nearest neighbours around a lattice cell; the longer range of interactions are not considered
 |
| |
|---|---|---|
| Figure 35: Plot of the reduced temperature against 1/(lattice length) | Figure 36: Code used to produce Figure 35, in addition to the extrapolation of the |
JC: Good agreement between Monte Carlo and analytical result. Is it worth including the 2x2 data in the fit as you have shown that this size lattice is not good at describing behaviour?
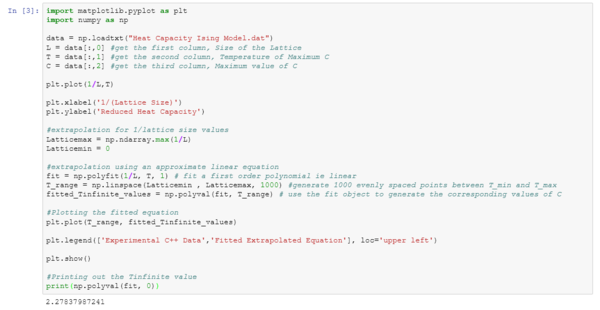
TASK: A plot that uses the scaling relation to determine
import matplotlib.pyplot as plt
import numpy as np
data = np.loadtxt("Heat Capacity Ising Model.dat")
L = data[:,0] #get the first column, Size of the Lattice
T = data[:,1] #get the second column, Temperature of Maximum C
C = data[:,2] #get the third column, Maximum value of C
plt.plot(1/L,T)
plt.xlabel('1/(Lattice Size)')
plt.ylabel('Reduced Heat Capacity')
#extrapolation for 1/lattice size values
Latticemax = np.ndarray.max(1/L)
Latticemin = 0
#extrapolation using an approximate linear equation
fit = np.polyfit(1/L, T, 1) # fit a first order polynomial ie linear
T_range = np.linspace(Latticemin , Latticemax, 1000) #generate 1000 evenly spaced points between T_min and T_max
fitted_Tinfinite_values = np.polyval(fit, T_range) # use the fit object to generate the corresponding values of C
#Plotting the fitted equation
plt.plot(T_range, fitted_Tinfinite_values)
plt.legend(['Experimental C++ Data','Fitted Extrapolated Equation'], loc='upper left')
plt.show()
#Printing out the Tinfinite value
print(np.polyval(fit, 0))
|
- ↑ Peter Atkins and Julio De Paula, Physical Chemistry, Oxford University Press, 5th edition, 2009, page 415
- ↑ Maginn EJ, Shah JK. Ising Lattice [Internet]. Department of Chemical and Biomolecular Engineering, University of Notre Dame; [cited 2016]. Available from: http://www.peq.coppe.ufrj.br/thermo/lecture_notes/ising.pdf
- ↑ 3.0 3.1 Onsager, L. Crystal Statistics I A Two-Dimensional Model with an Order-Disorder Transition. Phys Rev. [Online] 1944;65(3-4): 117-149. Available from: http://link.aps.org/doi/10.1103/PhysRev.65.117, doi :10.1103/PhysRev.65.117, [Accessed 15 November 2016].