Sns18
NH3 Molecule
General Information
| Molecule | NH3 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p.) |
| Final Energy E(RB3LYP) (au) | -56.55777 |
| RMS Gradient | 4.85 x 10-6 |
| Point Group | C3v |
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986260D-10
Optimization completed.
-- Stationary point found.
- N-H Bond distance: 1.02 Å
- H-N-H Bond angle: 106°
NH3 Molecule |
Vibrational Analysis
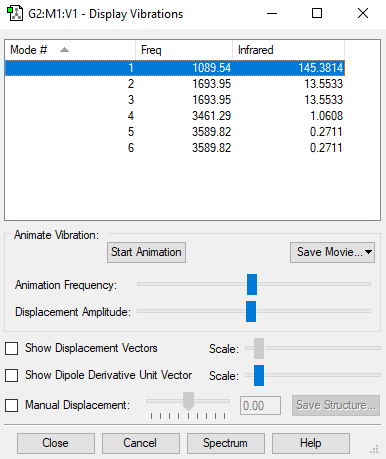
| Wavenumber / cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity / arbitrary units | 145 | 14 | 14 | 1 | 0 | 0 |
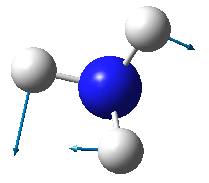
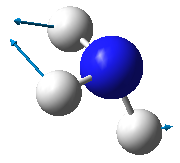
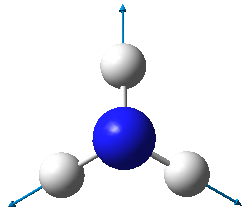
| Image | 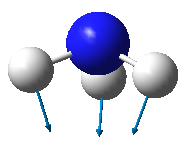
|
|
|
|
|
|
- From the 3N-6 rule, ammonia is expected to have 6 modes of vibration ((3x4)-6).
- The two vibrational modes with wavenumber 1694 cm-1 are degenerate; and the two modes with wavenumber 3590 cm-1 are degenerate.
- The vibrations with wavenumbers 1090 cm-1, 1694 cm-1, and 1694 cm-1 are bending vibrations. The vibrations with wavenumbers 3461 cm-1, 3590 cm-1, and 3590 cm-1 are bond stretches.
- The vibrational mode with wavenumber 3461 cm-1 is highly symmetric.
- The last mode with wavenumber 3590 cm-1 is the "umbrella" mode (the mode furthest right in the above table).
- Two bands would be expected in an experimental spectrum of gaseous ammonia.
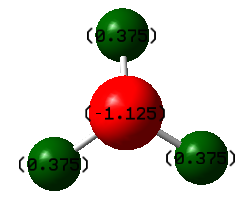
Atomic Charges
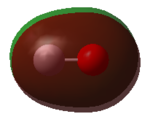
The N-atom has a -1.125 charge, and the three H-atoms each have a charge of 0.375. Nitrogen is more electronegative than hydrogen, so it is expected that the N-atom would have a slightly negative charge while the surrounding H-atoms have a slightly positive charge. The nitrogen pulls electron density away from the hydrogens.
N2 Molecule
General Information
| Molecule | N2 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p.) |
| Final Energy E(RB3LYP) (au) | -109.52413 |
| RMS Gradient | 6.0 x 10-7 |
| Point Group | D∞h |
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401050D-13
Optimization completed.
-- Stationary point found.
- N-N Bond distance: 1.11 Å
N2 Molecule |
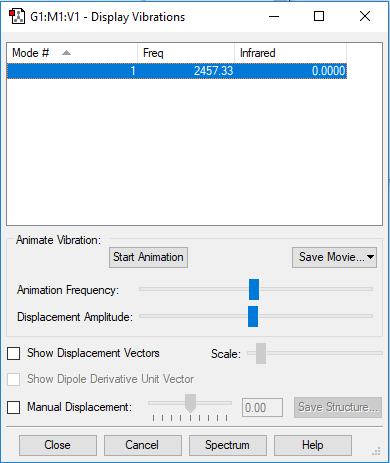
Vibrational Analysis
| Wavenumber / cm-1 | 2457 |
|---|---|
| Symmetry | SGG |
| Intensity / arbitrary units | 0 |
| Image | 
|
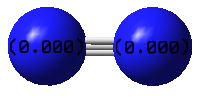
Atomic Charges
Both N-atoms have zero charge. They are of equal electronegativity so neither atom can pull electron density away from the other. If the image showed colour by charge, both atoms would be black, so they were left as blue so that the numbers could be viewed.
H2 Molecule
General Information
| Molecule | H2 |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p.) |
| Final Energy E(RB3LYP) (au) | -1.17854 |
| RMS Gradient | 1.7 x 10-7 |
| Point Group | D∞h |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
- H-H Bond distance: 0.74 Å
H2 Molecule |
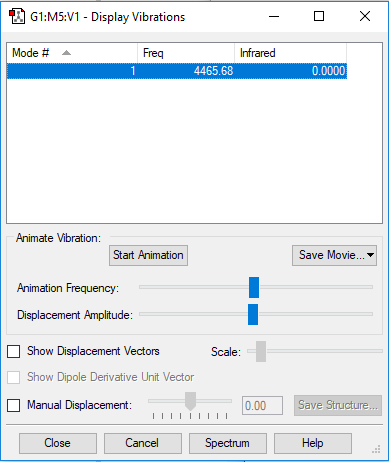
Vibrational Analysis
| Wavenumber / cm-1 | 4466 |
|---|---|
| Symmetry | SGG |
| Intensity / arbitrary units | 0 |
| Image | 
|
Atomic Charges
Both H-atoms have zero charge. They are of equal electronegativity so neither atom can pull electron density away from the other. If the image showed colour by charge, both atoms would be black, so they were left as white so that the numbers could be viewed.
Mono-metallic TM Complex
TRANS-BIS(1,1'-BIS(DIETHYLPHOSPHINO)FERROCENE)-bis(dinitrogen)-molybdenum is a mono-metallic TM complex that coordinates N2. The unique identifier is AGISIT. Link to the complex: [[1]]
The N-N bond distances in this complex are 1.12 Å[1] compared to 1.11 Å in the optimised nitrogen molecule. The bond distances are longer in the complex because the transition metal attracts the electron density away from the bond, causing the N-N bond to weaken, and thus become longer.
There will be differences between the computational and experimental values for bond distances because the calculation method used for the computational analysis is imperfect and will not be completely accurate. It does not take all possible parameters into account, so the values it produces will not be entirely reliable. Experimental values will have associated errors depending on the techniques used to measure the bond distances and the samples analysed.
Haber-Bosch Process
- E(NH3)= -56.55777 au
- 2*E(NH3)= -113.11554 au
- E(N2)= -109.52413 au
- E(H2)= -1.17854
- 3*E(H2)= -3.53562 au
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579 au
- ΔE= -146.5 kJ mol-1
The energy change for the conversion of hydrogen and nitrogen gas into ammonia is -146.5 kJ mol-1, so the reaction is exothermic. As a result, the ammonia product is more stable than the gaseous reactants, as energy is being released during the reaction.
CO Molecule
General Information
| Molecule | CO |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p.) |
| Final Energy E(RB3LYP) (au) | -113.30945 |
| RMS Gradient | 1.8 x 10-5 |
| Point Group | C∞v |
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000018 0.001200 YES
Predicted change in Energy=-3.956716D-10
Optimization completed.
-- Stationary point found.
C-O Bond distance: 1.14 Å
CO Molecule |
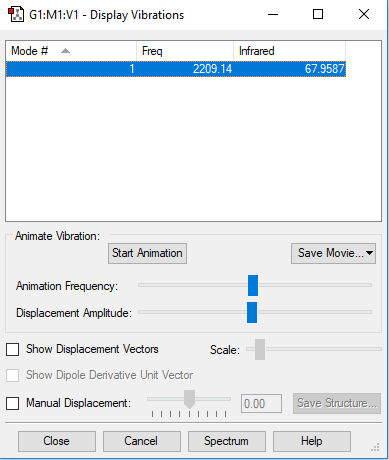
Vibrational Analysis
| Wavenumber / cm-1 | 2209 |
|---|---|
| Symmetry | SG |
| Intensity / arbitrary units | 68 |
| Image | 
|
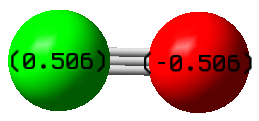
Atomic Charges
The oxygen atom has a partial negative charge and the carbon has a partial positive charge of the same magnitude. This is because the oxygen is more electronegative than the carbon so it attracts electron density away from the carbon atom, resulting in partial charges on both atoms.
Molecular Orbitals
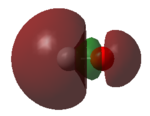
- Molecular orbital (MO) 1 is a 1π bonding orbital formed by the combination of two 2py atomic orbitals (AOs). There is also another degenerate occupied 1π MO that is perpendicular to MO 1.
- MO 2 is a 1σ bonding orbital. It is formed by the combination of two 1s orbitals but it is very deep in energy so there is almost no overlap between the two 1s orbitals of carbon and oxygen.
- MO 3 is the HOMO. It is an occupied 5σ bonding orbital, that results in the molecule’s σ bond between the carbon and the oxygen. MO 3 is formed from the overlap of two 2pz AOs.
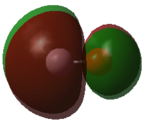
- MO 4 is a 1π* antibonding orbital formed by the combination of two 2py AOs that are out of phase. There is also another degenerate occupied 1π* MO that is perpendicular to MO 4.
- Because both the bonding 1π orbitals are occupied and both the antibonding 1π* orbitals are unoccupied, this results in CO having two π bonds. Therefore, CO has two π bonds and one σ bond, meaning that it has a triple bond between the atoms.
- MO 5 is a 6σ* antibonding orbital that is fairly high in energy. It is formed by the combination of two 2pz AOs that are out of phase with each other.
H2O Molecule (Independent)
General Information
| Molecule | H2O |
|---|---|
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p.) |
| Final Energy E(RB3LYP) (au) | -76.41974 |
| RMS Gradient | 6.28 x 10-5 |
| Point Group | C2v |
- O-H Bond distance: 0.97 Å
- H-O-H Bond angle: 104°
Item Value Threshold Converged?
Maximum Force 0.000099 0.000450 YES
RMS Force 0.000081 0.000300 YES
Maximum Displacement 0.000128 0.001800 YES
RMS Displacement 0.000120 0.001200 YES
Predicted change in Energy=-1.939669D-08
Optimization completed.
-- Stationary point found.
H2O Molecule |
Vibrational Analysis
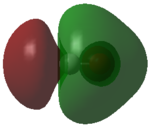
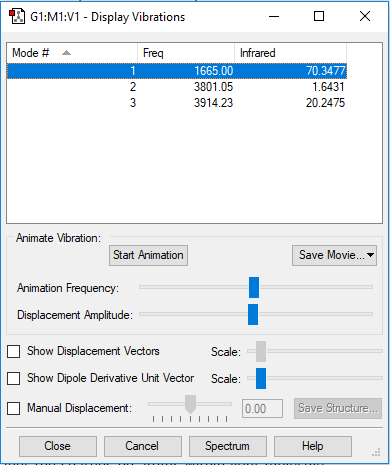
| Wavenumber / cm-1 | 1665 | 3801 | 3914 |
|---|---|---|---|
| Symmetry | A1 | A1 | B2 |
| Intensity / arbitrary units | 70 | 2 | 20 |
| Image | 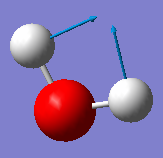
|
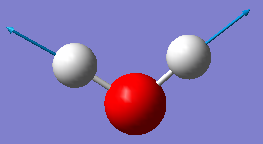
|
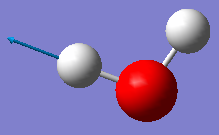
|
Atomic Charges
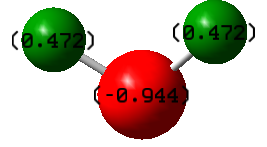
The oxygen atom is more electronegative than the hydrogen atoms, so electron density is drawn away from the hydrogen atoms. This causes the oxygen to have a partial negative charge and the hydrogen atoms to have equal partial positive charges. Due to the bent shape of the molecule, H2O has an overall dipole.
- ↑ (1)M.Yuki, Y.Miyake, Y.Nishibayashi, I.Wakiji, M.Hidai CCDC 704103: Experimental Crystal Structure Determination, 2009, DOI: 10.5517/ccrmp01.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - the mode at 1090cm-1 is the umbrella mode
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3.5/5
Have you completed the calculation and included all relevant information?
YES - you could have explained the calculated vibration.
Have you added information about MOs and charges on atoms?
YES
Only the 1s of O contributes to MO2. It is a non-bonding orbitals rather than a bonding one. You could have explained the energetic order of the MOs (number of nodes...).
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
YES - you could have explained the computed vibrations.
Do some deeper analysis on your results so far