Sn518
NH3 Molecule
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986275D-10
Optimization completed.
-- Stationary point found.
NH3 Molecule |
Molecule Summary
| Molecule | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy E(RB3LYP)(au) | -56.55777 |
| RMS Gradient(au) | 4.85x 10-6 |
| Point Group | C3v |
Bond Length: 1.02Å H-N-H Bond Angle: 105.7°
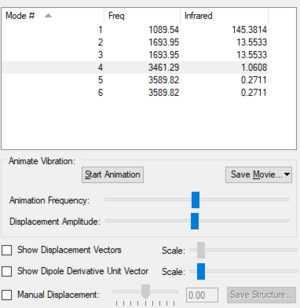
Vibrations
| Vibrational Mode | 1 | 2 | 3 | 4 | 5 | 6 |
| Wavenumber (cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity (arbitrary units) | 145 | 14 | 14 | 1 | 0 | 0 |
| Vibration Image | 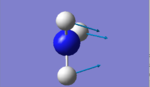 |
 |
 |
 |
 |
|
The expected number of vibrational modes for NH3 is 3N-6= 3(4)-6= 6. The degenerate modes are of wavenumber 1694 cm-1, and of wavenumber 3590cm-1. Modes with frequencies of 1090cm-1and the two modes of 1694cm-1 are bending modes whereas frequencies of 3461cm-1, and 3590 cm-1 are the bond stretching vibrations. The vibrational frequency of 3461cm-1with A1 symmetry is the highly symmetric mode. The umbrella mode appears to be the second vibrational mode (6) with wavenumber 3590cm-1. Two bands would be expected in an experimental spectrum of gaseous ammonia from the modes with frequency 1090cm-1 and 1694cm-1, but as two modes have peaks at 1696cm-1,they will overlap.
Charge Analysis
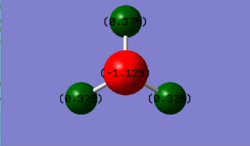
The nitrogen atom would be expected to be slightly negative as it is an electronegative atom whereas the H atom would be slightly positive. This correlates to the image shown above as the overall charge cancels out as a NH3 molecule does not possess any charge.
N2 Molecule
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400949D-13
Optimization completed.
-- Stationary point found.
N2 Molecule |
Molecule Summary
| Molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy E(RB3LYP)(au) | -109.52513 |
| RMS Gradient(au) | 6.00x 10-7 |
| Point Group | D∞h |
Bond Length: 1.11Å
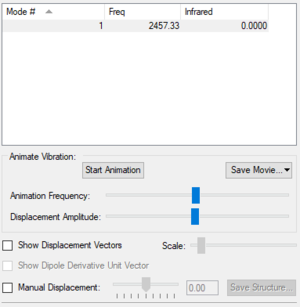
Vibrations
| Vibrational Mode | 1 |
| Wavenumber (cm-1) | 2457 |
| Symmetry | SGG |
| Intensity (arbitrary units) | 0 |
| Vibration Image | 
|
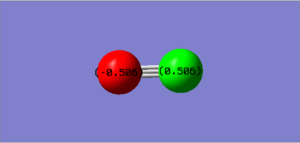
Charge Analysis
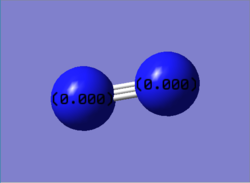
The charge is shown in the image as 0.00 as both nitrogen atoms have the same electronegativity, therefore the atoms could not be coloured by charge as a neutral charge is a black colour and the writing would be illegible as it is also black.
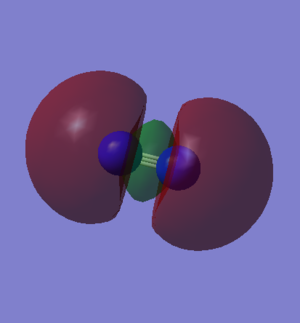
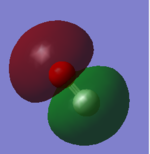
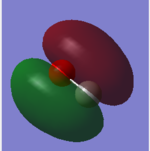
N2 Molecular Orbitals (Independence Mark)
HOMO is shown in the left image and is a 3σg orbital formed by contributions from the 2pz orbitals from each N atom in a N2 molecule. Whereas the LUMO is shown in the right image and is a 1π* orbital formed by contributions from 2px and 2py orbitals from both the N atoms.
H2 Molecule
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
H2 Molecule |
Molecule Summary
| Molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy E(RB3LYP)(au) | -1.17854 |
| RMS Gradient(au) | 1.7x 10-7 |
| Point Group | D∞h |
Bond Length: 0.74Å
Vibrations
Charge Analysis
Just like the N2 molecule, the charge is shown in the image as 0.00 as both hydrogen atoms have the same electronegativity. Thus, the neutral charge would be shown by the atoms being black.
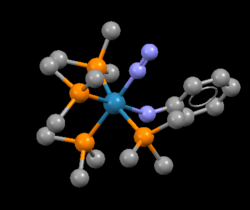
Mono-Metallic TM Complex: Phenylamido-(dinitrogen)-tetrakis(trimethylphosphine)-rhenium(i)
The structure found was Phenylamido-(dinitrogen)-tetrakis(trimethylphosphine)-rhenium(i) with the CSD refcode: BIBJAX. The N-N bond distance was 1.10Å.The crystal structure and N-N computational lengths are the same values to one decimal place which can be explained by the fact that there may not be much interaction between the transition metal and the nitrogen atoms. Nevertheless, it maybe the case that the errors are not reflected in the computational calculations and so both values appear to be identical.
Haber-Bosch Reaction Energy Calculation
E(NH3)=-56.55777 au
2*E(NH3)=-113.11554 au
E(N2)=-109.52413 au
E(H2)=-1.17854 au
3*E(H2)=-3.53562 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579au ΔE= -146.5 kJ mol-1
As the energy for this reaction: N2 + 3H2 -> 2NH3 is -146.5 kJ mol-1 and therefore negative, the reaction is an exothermic process and so the ammonia product is more stable than the gaseous product.
CO Molecule
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000018 0.001200 YES
Predicted change in Energy=-3.956716D-10
Optimization completed.
-- Stationary point found.
CO Molecule |
Molecule Summary
| Molecule | CO |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy E(RB3LYP)(au) | -113.30945 |
| RMS Gradient(au) | 1.83x 10-5 |
| Point Group | C∞v |
Bond Length: 1.14Å
Vibrations
| Vibrational Mode | 1 |
| Wavenumber (cm-1) | 2209 |
| Symmetry | SG |
| Intensity (arbitrary units) | 68 |
| Vibration Image | 
|
Charge Analysis
The charge on CO is neutral as the values of the charges cancel out.
CO Molecular Orbitals
All the orbitals here are occupied except the 1π* orbital which is the LUMO. In the CO molecule, the oxygen atom is more electronegative than the carbon atom and so the electrons are expected to occupy the orbitals that are predominantly of the oxygen atom. As oxygen is quite electronegative (3.4), its energy levels are deeper in energy (more negative) and so the system is more stable and the electrons are harder to remove. There are two 1π orbitals formed by the contribution of the 2px and 2py orbitals on C and O which are degenerate and there are in fact two 1π* antibonding orbitals again for the same reason. Therefore, in CO there are two π bonds and one σ bond. The 1π, 5σ and 1π* are a mixture of the 2p orbitals and thus the 5σ orbital is higher in energy than the 1π orbital. Both the HOMO and LUMO are not that different in terms of their energy values.
O2 Molecule (Independence Mark)
Item Value Threshold Converged?
Maximum Force 0.000130 0.000450 YES
RMS Force 0.000130 0.000300 YES
Maximum Displacement 0.000080 0.001800 YES
RMS Displacement 0.000113 0.001200 YES
Predicted change in Energy=-1.033738D-08
Optimization completed.
-- Stationary point found.
O Molecule |
Molecule Summary
| Molecule | O2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| Final Energy E(RB3LYP)(au) | -150.25742 |
| RMS Gradient(au) | 7.50x 10-5 |
| Point Group | D∞h |
Bond Length: 1.22Å
Vibrations
| Vibrational Mode | 1 |
| Wavenumber (cm-1) | 1643 |
| Symmetry | SGG |
| Intensity (arbitrary units) | 0 |
| Vibration Image | 
|
Charge Analysis
The two oxygen atoms have the same electronegativities and therefore pull the same amount of electron density. Thus the charge cancels out and the molecule is neutral.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, good explanations well done! You could have mentioned that the 1pi MO has a larger contribution from O than C, as you explained due to the different electronegativities causing the AOs of O to be lower than those of C. You can see the opposite effect in the LUMO.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You calculated O2 and you did deeper analysis on the MOs of N2, well done!