Resgrp:comp-photo-onthefly
Created and edited by S. Li.
The Direct (On-the-fly) CASSCF/RASSCF methods
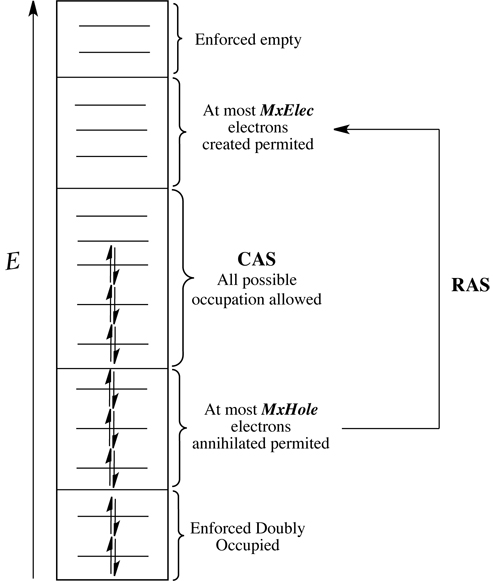
Complete active space self-consistent field (CASSCF) and restricted active space (RAS) SCF methods are powerful methods for describing all types of electronic structures, e.g. the ground states, excited states, positive and negative ions, etc, of chemical systems. A CASSCF calculation is a combination of a SCF calculation with a full CI in a window orbital space, which is called "the active space". Direct or "on-the-fly" means to recompute the Hamiltonian matrix elements whenever they are needed. Thus the huge memory requirement for storing these elements is avoided and systems with large active space are possible to be calculated by these methods. In Gaussian, this method requests the keyword CASSCF(N,M) to start such a type of calculation, where N indicates the number of electrons and M gives the number of MOs in the active space (N <= 2M). In order to obtain qualitative CASSCF/RASSCF calculation results on certain system, the selection of the active space for this system becomes crucial. The CASSCF tutorials suggest how this active space selection for certain system is carried out as well as how to run a CASSCF calculation. The RASSCF method divides the CAS active space, M, into three subset: RAS1, RAS2 and RAS3. RAS1 subspace is consistent of all doubly occupied orbitals, at most MxHole electrons can be annihilated from this subspace. RAS3 subspace contains all empty orbitals and at most MxElec electrons can be created in this subspace. There is no occupation restriction in the RAS2 subspace (see picture below).
In order to run a RASSCF job, the keyword: CASSCF(N,M,RASSCF(a,b,c,d)) must be used, where a = MxHole, b gives the number of orbitals contained in the RAS1 subspace, c = MxElec, and d indicates the number of orbitals in RAS3. A detailed explanation of the keyword: CASSCF (including the keyword of RASSCF) can be found on the Gaussian online manual [see web page].
The New Direct CASSCF/RASSCF Codes
The new direct CAS/RAS code introduces a matrix multiplication scheme based on the direct Hamiltonian matrix elements generation scheme of the current method [Klene2000 and Klene2003](G09 version). Thus highly-optimized linear algebra routines, which are able to use computer hardware efficiently, can be used in the new code, resulting in a much better performance overall than the current method. The side effect of this new method is that it requires some extra memory to store the additional intermediate matrices. For CASSCF, 4 (usual calculations, e.g. singlet calculation) or 5 (Slater determinant basis calculation) times of the square of the number of alpha strings (obtained from the output of l405 before gaussian enters l510) words of extra memory will be needed. For RASSCF, 4 or 5 times of the number of determinants (also obtained from the output of l405, note: if a default RAS singlet calculation or a Hartree-Waller triplet function basis calculation* is carried out, the number of determinants is about two times of the number of configurations obtained from l405) words of extra memory will be required.
* A default singlet RAS calculation uses Hartree-Waller singlet function basis (equivalent to set iop(4/46=1,5/39=1) explicitly). A Hartree-Waller triplet function basis RAS calculation is turned on by setting iop(4/46=2,5/39=2) explicitly. A default triplet calculation uses Slater determinant basis.
New features of the new code
1. The new code performs much faster than the current one. And when the active space (for CAS, if RAS, the size of RAS2 subspace) grows larger, the new code can perform faster and faster (one example is given in the table and picture* below).
System Number of Configurations CPU Time Used by l510 (s) New Code Speedup
Naphthalene
CAS(10,10) 63504 New: 100.9 1.50
Slater Det.serial Old: 151.1
Acenaphthylene
CAS(12,12) 853776 New: 2528.7 3.61
Slater Det.serial Old: 9132.3
Pyracylene
CAS(14,14) 11778624 New: 32928.5 4.83
Slater Det.serial Old: 158973.1
* These are single point energy calculations carried out by using Intel Nehalem servers (2 Intel Nehalem Quad-core 2.5 GHz processors per node with a peak speed of ~80 GFlops.
2. For shared-memory parallelism (CASSCF only), apart from the existing parallel scheme in the current code, the new code introduces a parallel scheme on the level of matrix multiplication. Thus the new code can be carried out via shared memory parallelism in two levels: on UPPER level (the existing parallel scheme), on LOWER level (parallel on matrix multiplication) or a combination of both. At UPPER LEVEL, the job is broken into many small tasks so these tasks can be calculated simultaneously. Each task has its own matrix multiplication to carry out. Thus at UPPER LEVEL, the matrix multiplication is carried out serially for each task. By contrast, at LOWER LEVEL, the job is still broken into many small tasks, but all the processors will carry out one task a time with the corresponding matrix multiplication operated in parallel using the available processors. This introduces more flexibility of running parallel CASSCF jobs especially when the available memory is not large enough. This is because the existing shared memory parallel scheme requires more memory when the number of shared memory processors increases. When the available memory is limited, this feature of the current code will restrict the shared memory parallel applications. By contrast, when using the new code and parallel on the Lower level, no matter how many shared memory processors are used, the required memory will remain constant.
Access to the new code
The newly developed direct CAS/RAS-SCF codes are available under the gaussian development version h01 (the current available development versions of gaussian can be found here). In order to access to this version of gaussian, one needs to load it first by typing (on cx1):
-bash-3.2$ module load gaussian/devel-modules -bash-3.2$ module load gdvh01_725
After doing this, by typing
-bash-3.2$ which gdv
one should be able to check whether the correct gaussian version is loaded as
/home/gaussian-devel/gaussiandvh01_pgi_725/gdv/gdv
After the right version of gaussian is loaded, in order to USE the new code, one needs to carry out the following steps:
1. use the %subst command in the LINK0 part of the gaussian input file to take the executable l510 from the given directory. The directory is slightly different when using shared memory parallelism from using distributed memory parallelism (more detail about this will be given below).
2. add iop(5/139 = n) (for CASSCF jobs, n >= 1; for RASSCF jobs, n = 1 only) explicitly in the Route section of the gaussian input file. Otherwise the current CAS/RAS code will be used. The reason for setting n >= 1 for CASSCF jobs rather than setting n = 1 only is because when using CASSCF, there are two levels of shared memory parallelism. The value of n in CASSCF controls what level of shared memory parallelim is used in a CASSCF calculation (see example below).
In order to use the new code for serial or shared-memory parallel CASSCF / RASSCF jobs, FIRSTLY, the following lines must be added in the LINK0 part of the input file:
1. if your job runs serially, then
%subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras %NProcShared=1
or
%subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras
is added
2. if your job runs parallel using shared-memory, e.g. using 4 cores of certain cx1 node, then
%subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras %NProcShared=4
is added. THEN in the Route section of the input file, one needs to add iop(5/139 = n) explicitly.
For example, if one carries out a CASSCF(10,10) calculation on naphthalene using 4 processors for shared-memory parallelism, the route section of the input file will look like:
#p casscf(10,10)/4-31g guess=(read,alter) geom=check iop(5/139=1)
And the entire input file for this example is given as:
%nosave %chk=CAS10_test #p HF/4-31g pop=full nosymm // this step is for selecting the right orbitals in the active space (see CASSCF tutorials ) Naphthalene HF Linkto CASSCF singlet full calculation. 0 1 C -2.42610388 -0.70881491 0.00000000 C -1.24119954 -1.40205677 0.00000000 C -0.00000000 -0.70918612 -0.00000000 C -0.00000000 0.70918612 -0.00000000 C -1.24119954 1.40205677 -0.00000000 C -2.42610388 0.70881491 -0.00000000 H 1.23306389 -2.50189963 0.00000000 H -3.38797619 -1.24247368 0.00000000 H -1.23306389 -2.50189963 0.00000000 C 1.24119954 -1.40205677 -0.00000000 C 1.24119954 1.40205677 0.00000000 H -1.23306389 2.50189963 0.00000000 H -3.38797619 1.24247368 -0.00000000 C 2.42610388 0.70881491 0.00000000 C 2.42610388 -0.70881491 -0.00000000 H 1.23306389 2.50189963 -0.00000000 H 3.38797619 1.24247368 0.00000000 H 3.38797619 -1.24247368 -0.00000000 --Link1-- %subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras %NProcShared=4 %nosave %chk=CAS10_test #p casscf(10,10)/4-31g guess=(read,alter) geom=check iop(5/139=1) nosymm Naphthalene CASSCF(10,10)/4-31G orbital switch according to HF calculation 0 1 27 30 //switch the right orbitals for the active space 38 41 39 47
If one runs a RASSCF job, the process is similar, and the RASSCF input file (part) will read as:
... %subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras %NProcShared=4 %nosave %chk=RAS10_2222_test #p casscf(10,10,rasscf(2,2,2,2))/4-31g guess=(read,alter) geom=check iop(5/139=1) nosymm //for this type of jobs, iop(5/139=1) only ...
Once the CAS/RAS input file is set up, one can carry out the calculation by typing (assuming the name of the input file is a.com / a.gjf):
-bash-3.2$ gdv a.com &
This will generate the output file in the same directory where the input file is in. In order to generate the output file in different directory, the path of the directory must be set explicitly. Examples of doing this can be found in the Examples section below.
As mentioned above, the new CASSCF code has two levels of shared-memory parallelism. One is called the Upper Level, which is the parallel scheme used in the current code. The other is called the Lower Level that parallelizes the program on the level of matrix multiplication.
When one sets
%NProcShared = X
explicitly to use shared-memory parallelism for certain CASSCF job, s/he can choose to use upper level parallel, lower level parallel or a combination of both by varying the value n of iop(5/139 = n).
1. To turn on UPPER LEVEL parallelism, one only needs set
iop(5/139 = 1)
in the route section of the input file.
2. To turn on LOWER LEVEL parallelism, the value becomes
iop(5/139 = X)
3. In order to use the combination of both, the value of iop(5/139) must be set as an integer smaller than X but greater than 1 (e.g. 1 < n < X). Then the upper level tasks will be distributed to int(X/n) processors and the matrix multiplications corresponding to each task will be parallelized by n processors.
For example, if one sets %NProcShared = 8 in the LINK0 part and iop(5/139 = 1) in the route section for certain CAS job, according to above description, ONLY Upper Level parallelism will be used to carry out this job. However, when the user sets iop(5/139 = 8) in the route section for the job, then ONLY Lower Level parallelism will be used. Alternatively, if the user sets the value of iop(5/139) less than 8 but greater than 1, e.g. iop(5/139 = 4) then 2 processors will be used for UPPER LEVEL parallelism and 4 processors will be used for LOWER LEVEL parallelism. This combination is a simulation of using Linda via shared-memory parallelism.
---Known Problems---
Although Lower level shared memory parallel for CASSCF jobs doesn't require extra memory when the number of processors increases, the scaling of the parallel at this level is very poor. For example, if one sets %NProcShared=8, the scaling of the parallelization on matrix multiplication can only reach 3 on top. By contrast, the scaling of parallelization on Upper Level can reach 6 or 7 depending on the type of hardware one uses, e.g. the Nehalem processors. This might be because of the parallel overhead when carrying out the parallel at lower level. However, this has not been explicitly tested. So currently the real reason for this is still unknown.
Thus when the available memory is enough, one is suggested to use the UPPER LEVEL shared-memory parallelism to run CASSCF jobs (iop(5/139 = 1)).
Running distributed-memory parallel jobs (Linda) with the new code
In order to run a CAS/RAS job in distributed memory parallelism (Linda) or a combination of shared memory parallelism and distributed memory parallelism using the new code, in the LINK0 part of the input file, one needs to add:
%subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras/linda-exe %NProcLinda = X %NProcShared = Y
where the value of X gives the number of nodes one requires and the value of Y indicates the number of shared-memory processors on each node one requires. Thus in total, there will be X*Y processors used for parallelism. The values of X and Y can be:
- X = 1, Y = 1 : Serial job only (see above description)
- X = 1, Y > 1 : Shared-memory parallelism only (see above)
- X > 1, Y = 1 : Distributed-memory parallelism only (this is seldom used in practice nowadays as modern computer architectures include multi-core parallel processors)
- X > 1, Y > 1 : A combination of shared-memory and distributed memory parallelism.
THEN in the Route Section of the input file, the iop(5/139 = 1) (set the value as 1 to achieve the best parallel scaling) is added. Still using the above naphthalene example, the input file of a parallel job using 2 nodes with 4 cores per node (thus in total 8 cores will be used for the job) will look like:
%nosave %chk=CAS10_Linda_test #p HF/4-31g pop=full nosymm Naphthalene HF Linkto CASSCF singlet single point energy calculation. 0 1 THE COORDINATE OF ATOMS --Link1-- %subst l510 /home/gaussian-devel/gaussiandvh01_pgi_725/shaopeng_casras/linda-exe %NProcLinda=2 %NProcShared=4 %nosave %chk=CAS10_Linda_test #p casscf(10,10)/4-31g guess=(read,alter) geom=check iop(5/139=1) nosymm (if RAS job, this line becomes) #p casscf(10,10,rasscf(2,2,2,2))/4-31g guess=(read,alter) geom=check iop(5/139=1) nosymm Naphthalene CASSCF(10,10)/4-31G Linda calculation with orbital-switch according to HF calculation 0 1 27 30 38 41 39 47
In order to carry out this Linda parallel calculation, the command
gdvlb
will be required rather than gdv.
Some examples
As how to use the new code has been described above, in this section several examples of test jobs will be provided. In order to get the result quick, all the examples are single point energy calculations of two systems with relatively small active space, e.g. 10 active orbitals for CAS serial demonstration and 12 active orbitals for CAS parallel demonstration. Naphthalene is used in the 10 active orbitals examples and acenaphthylene is used in the 12 active orbitals examples. For RAS demonstration, the active space is set larger, e.g. calculations on acenaphthylene with a large active space (20 orbitals: 4 in RAS1, 4 in RAS2, and 12 in RAS3 with double excitation) are the serial demonstrations and calculations on pyracylene with the active space of 20 orbitals (4 in RAS1, 6 in RAS2, and 10 in RAS3 with double excitation) are used for parallel demonstrations.
Serial calculation
CASSCF test job
The input file of a CASSCF(10,10) job on naphthalene can be found: Media:CAS10in10_New_code_serial.gjf
The corresponding job submit script reads as:
#PBS -l select=1:ncpus=1:mem=7200mb:nehalem=true //This line tells the cluster to use 1 processor on 1 node and 7200MBytes memory for this job. And this job uses Nehalem processors only #PBS -l walltime=72:00:00 #PBS -joe module load gaussian/devel-modules module load gdvh01_725 gdv < [Input file path]/CAS10in10_New_code_serial.gjf > [Output file path]/CAS10in10_New_code_serial_test.log
Note: if the queue, e.g. "q48" or "pqmb", one uses on cx1 doesn't include nodes with Nehalem processors, which give better scaling when a program is parallelized compared to other available processor types (except those newer ones) on cx1, this job won't be carried out. So normally the "nehalem=true" won't be necessary in the jobscript. But pqchem has a few nodes with Nehalem processors. When using this queue on cx1, it is better to add this command in the job submitting script.
The corresponding output file: Media:CAS10in10_New_code_serial_test.log
To compare the performance to the new code, the same job of using current code is carried out too:
Media:CAS10in10_Current_code_serial.gjf
and
Media:CAS10in10_Current_code_serial_test.log
RASSCF test job
For the serial RASSCF example, as described above, a RAS(12,4+4+12)[2,2] calculation is carried out on acenaphthylene.
The input file is: Media:RAS12in12_24212_New_code_serial.gjf
And the output file is: Media:RAS12in12_24212_New_code_serial_updated_test.log
The same job is also carried out using the current code:
the input file: Media:RAS12in12_24212_Current_code_serial.gjf
the output file: Media:RAS12in12_24212_Current_code_serial_updated_test.log
Upper level parallel example
CASSCF test jobs
Parallel calculation on acenaphthylene using 4 shared memory processors via the new code:
Media:CAS12in12_New_code_shared_4P.gjf
The corresponding jobscript reads
#PBS -l select=1:ncpus=4:mem=7200mb #PBS -l walltime=72:00:00 #PBS -joe module load gaussian/devel-modules module load gdvh01_725 gdv < [input file path]/CAS12in12_New_code_shared_4P.gif > [output file path]/CAS12in12_New_code_shared_4P_test.log
Media:CAS12in12_New_code_shared_4P_test.log
The same job using the current code:
Media:CAS12in12_Current_code_shared_4P.gjf and
Media:CAS12in12_Current_code_shared_4P_test.log
RASSCF test jobs
A RAS(14,4+6+10)[2,2] calculation is carried out on pyracylene using shared-memory parallelism.
The input file : Media:RAS14in14_24210_New_code_shared_8P.gjf
and the output file : Media:RAS14in14_24210_New_code_shared_8P_test.log
The same job is carried out using the current code as well:
input file : Media:RAS14in14_24210_Current_code_shared_8P.gjf
output file : Media:RAS14in14_24210_Current_code_shared_8P_test.log
Lower level parallel (CAS only) example
Although the scaling of the lower level parallel is not as good as the scaling of the upper level parallel, one example of using the lower level parallel must be given.
The input file : Media:CAS12in12_New_code_shared_lower_4P.gjf
Note: in this input file, the value of iop(5/139) is set the same as the value of %NProcShared = 4 (more detail see above description).
And the output file : Media:CAS12in12_New_code_shared_lower_4P_test.log
The number of shared-memory processors on a single PC node is limited. In order to save more time when run a program in parallel, one usually uses a combination of distributed-memory parallelism and shared-memory parallelism. This allows one to use more processors to carry out the job. Thus more tasks can be carried out at the same time. How to use this combination has been described above. Here only gives one example of such calculation for CAS and one for RAS respectively.
CASSCF Linda job
CASSCF calculation on acenaphthylene using 2 nodes with 4 processors per node (thus in total 8 processors are used):
input file: File:CAS12in12 New code Linda 2N4P.gjf
the corresponding job submitting script reads as:
#PBS -l select=2:ncpus=4:mem=15GB* //this requires 2 nodes, 4 processors and 15GBytes per node. #PBS -l walltime=72:00:00 #PBS -joe module load gaussian/devel-modules module load gdvh01_725 gdvlb < [input file path]/CAS12in12_New_code_Linda_2N4P.com > [output file path]/CAS12in12_New_code_Linda_2N4P_test.log
* the reason for setting "mem=15GB", which is more than enough for the job, for this job is because if the amount of memory on one node is greater than the select*mem (the multiplication between the value of "select" and the value of "mem") and the number of shared memory processors on one node is greater or equal to select*ncpus, this Linda job will be carried out on one physical node rather than two nodes. In this case, the memory of the node is "partitioned" into two parts. 4 processors share one part and the other 4 share the other part (this can also be achieved by setting "select=1:ncpus=8:mem=8GB" explicitly). Therefore, in order to use two physical nodes to carry out this job, one can either require a large amount of memory on each node or set "ncpus" close to the number of total processors one node has.
the output file for this job is:
Media:CAS12in12_New_code_Linda_2N4P_test.log
The same job calculated using the current code:
input file: Media:CAS12in12_Current_code_Linda_2N4P.gjf
output file: Media:CAS12in12_Current_code_Linda_2N4P_test.log
RASSCF Linda job
RASSCF(14,4+6+10)[2,2] singlet calculation on pyracylene using 2 nodes 4 processors per node:
Using the new code:
input file: Media:RAS14in14_24210_New_code_Linda_2N4P.gjf
output file: Media:RAS14in14_24210_New_code_Linda_2N4P_test.log
Using the current code:
input file: Media:RAS14in14_24210_Current_code_Linda_2N4P.gjf
output file: Media:RAS14in14_24210_Current_code_Linda_2N4P_test.log