Rep:Title=Mod:ChlorineDJN16
ammonia
optimisation
optimised information
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -56.55776873 a.u.
RMS Gradient Norm = 0.00000485 a.u.
Point Group = C3V
bond length (N-H) = 1.01798Å
bond angle (H-N-H) = 105.741°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986273D-10
Optimization completed.
-- Stationary point found.
optimised NH3
optimised NH3 |
optimisation file for NH3 is linked here
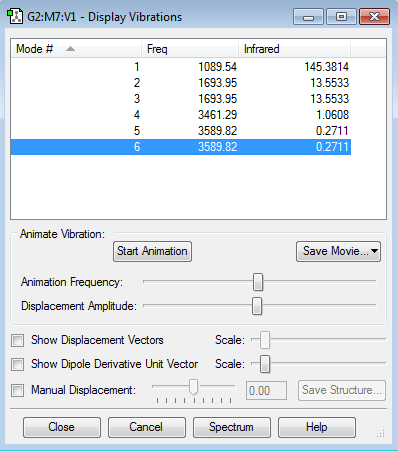
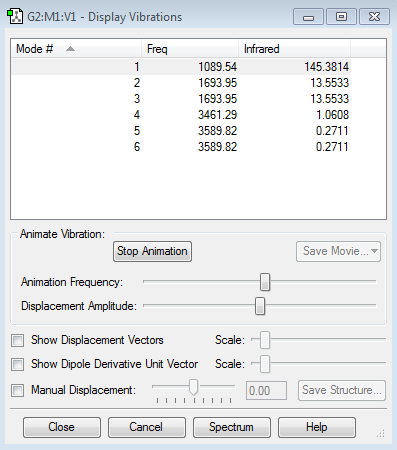
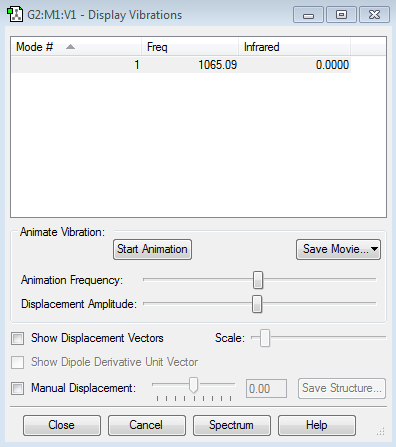
Vibrational modes
Questions:
1. How many modes of vibration are expected from the 3N-6 rule?
2. Which modes are degenerate?
3. Which modes are bending? which are stretching?
4. Which modes are highly symmetric?
5. Which mode is described as 'umbrella' mode?
6. How many main peaks should you expect to see in a spectrum for gaseous ammonia?
Ammonia can absorb IR because it has a dipole moment. Ammonia should show six vibrational modes. The bends 2&3 are degenerate and the stretches 5&6 are degenerate because the movements are symmetrical but the protons that are bending/stretching are different. Mode 4 is highly symmetric. Mode 1 is 'umbrella'. The two main peaks in the spectrum are due to mode 1 modes 2/3 (there are also two minuscule peaks due to modes 4 and 5/6)
| number of mode | description |
|---|---|
| 1 | umbrella, symmetric mode, bend |
| 2 | bend |
| 3 | bend |
| 4 | symmetric, stretch |
| 5 | stretch |
| 6 | stretch |
Charge distribution
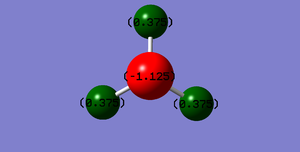
Nitrogen
optimisation
optimised information
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -109.52412868
RMS Gradient Norm = 0.00000060 a.u.
Point Group = D*H
bond length (N-N) = 1.10550Å
bond angle (N-N) = 180°
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.383662D-13
Optimization completed.
-- Stationary point found.
optimised N2
There are no IR peaks for N2 because there is no dipole moment.
optimised N2 |
optimisation file for N2 is linked here

Hydrogen
optimisation
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -1.17853936 a.u.
RMS Gradient Norm = 0.00000017 a.u.
Point Group = D*H
bond length (H-H) = 0.74279Å
bond angle (H-H) = 180°
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.167770D-13
Optimization completed.
-- Stationary point found.
There are no IR peaks for H2 because there is no dipole moment.
optimised H2
optimised H2 |

optimisation file for H2 is linked here
Haber-Bosch process
N2 + 3H2 -> 2NH3
E(NH3)= -56.55776873 a.u. 2*E(NH3)= -113.1155375 a.u. E(N2)= -109.52412868 a.u. E(H2)= -1.17853936 a.u. 3*E(H2)= -3.53561808 a.u. ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 a.u. ΔE= -146.478599028kJ/mol
literature value for haber bosch process: ΔH = -45.7kJ/mol. [1]
The value of ΔE for the computed values is lower than the literature value
Fluorine
optimised information
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -199.49825218 a.u.
RMS Gradient Norm = 0.00007365 a.u.
Point Group = D*H
bond length (F-F) = 1.40281Å
bond angle (F-F) = 180°
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995024D-08
Optimization completed.
-- Stationary point found.
optimised F2 |
optimisation file for F2 is linked here
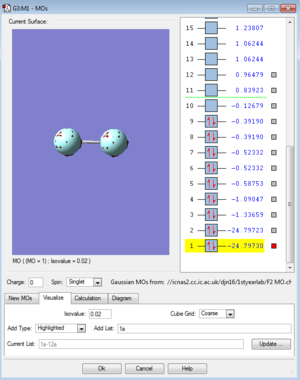
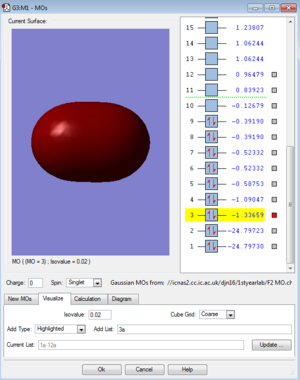
MOs for F2

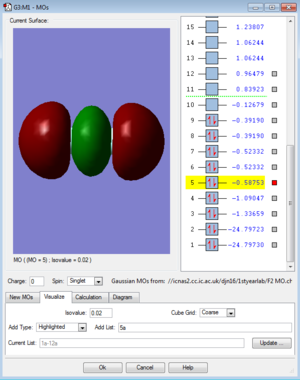
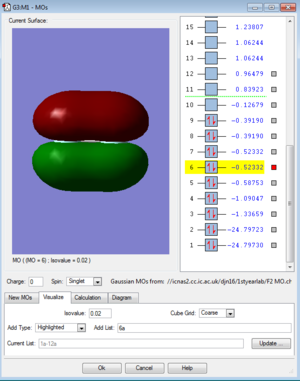
References
<references> [1]