Rep:Module2:Dima1407
TlBr3 optimisation
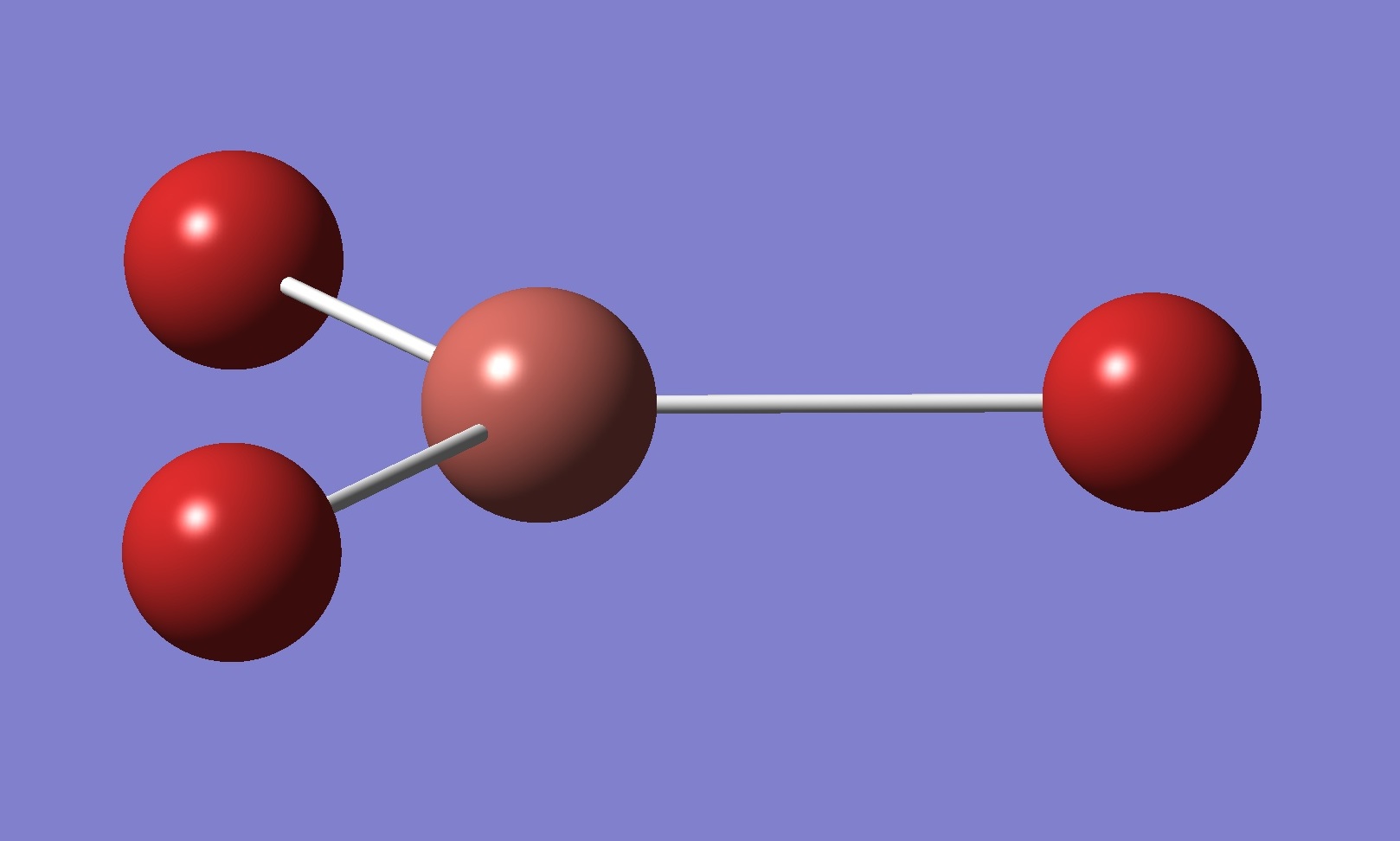
Bond length = 2.65 angstroms
Angle = 120,0 degrees
Molecular Orbitals and Vibrations of BH3
Completed population analysis for BH3 - DOI:10042/to-5800
| No. | Form of vibration | frequency | intencity | symmetry D3h point group | No. | Form of vibration | frequency | intencity | symmetry D3h point group | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
|
1150 | 93 | A2" | 4 |
|
2590 | 0 | A1' | |||||||
| 2 |
|
1200 | 12 | E' | 5 |
|
2730 | 104 | E' | |||||||
| 3 |
|
1200 | 12 | E' | 6 |
|
2730 | 104 | E' |
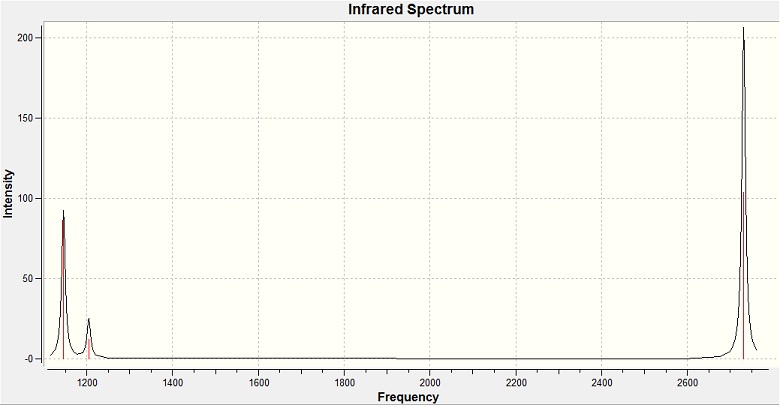
In our IR we have 3 peaks instead of 6. It is due to 2 peaks correspond to double degenerate vibration energy levels and intensity of fourth peak is 0, so we can't see it.
Calculations on TlBr3
Calculation method - RB3LYP
Basis set - LANL2DZ
We have to use the same method and basis set for both calculations because we want to do frequency analysis for a certain calculation we used. So if we used one method and basis set - we should use the same once in frequency analysis.
Because gradient could be zero in case of a maximum as well as in case of minimum, we need an additional test - frequency analysis, which is actually second derivative of potential energy surface.
| Low frequencies | -3.4226 | -0.0026 | -0.0004 | 0.0015 | 3.9361 | 3.9361 |
| Low frequencies | 46.4288 | 46.4291 | 52.1449 | 165.2685 | 210.6948 | 210.6949 |
As we can see all normal mode vibrations are positive, so optimised molecula is in the ground state (in its lowest energy level)
| Frequencies | 46 cm-1 | 46 cm-1 | 52 cm-1 | 170 cm-1 | 210 cm-1 | 210 cm-1 |
| Intensity | 4 | 4 | 6 | 0 | 25 | 25 |
Tl-Br bond length calculated with Gaussian is 2.65 angstroms, which is in a quite good agreement with literature value - 2.52[1].
GaussViw uses average bond length for all structures. So for example the bond length for PtCl2 and PtCl4 is the same. Sometimes when bond is bigger than an average bond, GaussiView does not draw in the bonds where we expect.
The MO diagram of BH3
| "real" MO | "LCAO" MO | |
|---|---|---|
| LUMO |  |
|
| HOMO |  |
|
| HOMO-1 | 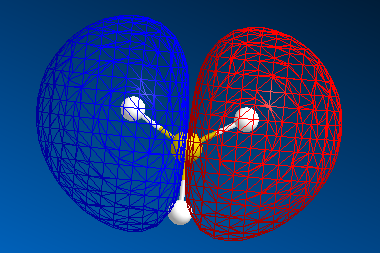 |

|
| HOMO-2 |  |

|
As we can see from the table the difference between "real" and "LCAO" MOs is quantitative - "real" MO can tell us energy and exact shape. But if we need relative energy levels and approximative shapes - LCAO is much better due to its simplicity.
Isomers of Mo(CO)4L2
Optimised trans-isomer of Mo(CO)4(PCl3)2 - DOI:10042/to-5928
Optimised cis-isomer of Mo(CO)4(PCl3)2 - DOI:10042/to-6107
| Vibration |
|
|
|
|
| |||||||||||||||
| Calculated values | freq. = 483cm-1, Intens. = 251 | freq. = 1938cm-1, Intens. = 1605 | freq. = 1942cm-1, Intens. = 813 | freq. = 1952cm-1, Intens. = 588 | freq. = 2019cm-1, Intens. = 545 | |||||||||||||||
| Literature value[2] | 1986cm-1 | 2004cm-1 | 2004cm-1 | 2072cm-1 |
| Vibration |
|
|
| |||||||||
| Calculated value | freq. = 435cm-1, Intens. = 801 | freq. = 1940cm-1, Intens. = 1606 | freq. = 1940cm-1, Intens. = 1606 | |||||||||
| Literature value | 1986cm-1 | 1986cm-1 |
| Bond | Mo-P trans | Mo-P cis | Mo-C trans | Mo-C cis1 | Mo-C cis12 | C-O |
|---|---|---|---|---|---|---|
| Calculated value, trans | 2.42 ang. | 2.48 ang. | 2.06 ang. | 2.05 ang. | 2.02 ang. | 1.17 ang. |
| Literature value[3][4] | 2.49* ang. | 2.49* ang. | 2.02-2.06 ang. | 1.98 ang. | 2.02 ang | 1.15 ang. |
The total energy of the cis- and trans-isomer are -623.69291203 a.u. and -623.69415610 a.u. The energy difference is 0.00124407 a.u. or 3.3 kJ/mol. Trans-isomer is more stable because bulky PCl3 groups are surrounded by small CO groups, but in case of cis-isomer two bulky PCl3 groups are near each other and this causes repulsion, which effects angles between the bonds (increase bending energy). Trans-isomer is also destabilized due to one P-Cl in PCl3 is eclipsed with C=O bond which causes angle C-Mo-P to differ from 90 degrees (), that is why energy difference etween two isomers are so small.
Doing our frequency test we did not get any negative values for both isomers, so we get ground state structures. Comparing carbonyl stretches obtained from the GaussView and found in literature we can say that values we have get are very close.
Also we have good results for bond lengths comparing to the literature values, so we can assume that our method is good and GaussView could be used for studding such a structures.
Mini-project: Complex compounds geometry
Complex compound with coordination number 4 could have 2 different geometries: square planar and tetrahedral. We are going to investigate using GaussView which one will Be lover In energy for Pt(NH3)Cl2 and Co(NH3)Cl2.
Completed optimisation for Pt(NH3)Cl2 - DOI:10042/to-6143
Completed optimisation for Co(NH3)Cl2 - DOI:10042/to-6142
Pt(NH3)Cl2 has a square planar structure which is as predicted for Pt compounds with d8 electron structure. Trans isomer is more stable due to its symmetry, but difference is negligible. Co complex has tetrahedral geometry[5]
Ion such As [Ni(CN)5]3- could have 2 geometric isomers: square pyramidal and trigonal bipyramidal. As we can see from the optimised files square pyramidal structure is more stable which is in accord with literature[6].
| Compound | Ni-C | Ni-C | Ni-C | C-O |
|---|---|---|---|---|
| Square pyr. | 1.87 | 1.90 | 1.83 | 1.19 |
| Trigonal bipyr. | 2.06 | 2.06 | 2.06 | 1.15 |
| Lit. sq. pyr. | 1.86 | 2.17 | 1.86 | |
| Lit. tr. bipyr. | 1.91 | 1.99 | 1.84 |
Completed optimisation for Square pyramidal - DOI:10042/to-6145
Completed optimisation for trigonal bipyramidal - DOI:10042/to-6144
From two examples we have viewed it is clear that GaussView is very powerful program to predict the geometry and MO for Complex compounds.
Reference
- ↑ J. Blixt; J. Am. Chem. Soc., 1995, 117 (18), pp 5089–5104 DOI:10.1021/ja00123a011
- ↑ Wai-Kee Li, Gong-du Zhou; Advanced structural inorganic chemistry
- ↑ Shishir Ghosh, Shariff E. Kabir; J. of Chem. Crystallography, Vol 40, N. 8,pp. 712-715 DOI:10.1007/s10870-010-9777-8
- ↑ CRC Handbook of Chemistry & Physics Online, 91st Edition, 2010-2011
- ↑ Dr. M. Pilkington; Principles of Inorganic Chemistry, Lecture 6
- ↑ Thomas G. Spiro; Inorg. Chem., 1970, 9 (11), pp 2415–242 DOI:10.1021/ic50093a006 10.1021/ic50093a006
