Rep:Module1pl506
Moduel 1
Exercise 1: The Hydrogenation of Cyclopentadiene Dimer
| cyclopentadiene dimer 1 | cyclopentadiene dimer 2 | ||||||
|---|---|---|---|---|---|---|---|
|
|
The two molecules shown above where drawn of Chem Bio 3D and the geometory was minimised using the MM2 structual calculation function of Chem Bio 3D. Below is a table with the data produced from these calculations.
| output data from MM2 calculations | dimer 1 | dimer 2 |
|---|---|---|
| Stretch | 1.2722 | 1.2631 |
| Bend | 20.5817 | 20.8691 |
| Stretch-Bend | -0.8330 | -0.8365 |
| Torsion | 7.6734 | 9.5014 |
| Non-1,4 VDW | -1.4353 | -1.5075 |
| 1,4 VDW | 4.2454 | 4.2851 |
| Dipole/Dipole | 0.3779 | 0.4443 |
| Total Energy (Kcal/mol) | 31.8823 | 34.0190 |
The data in the table shows that the reaction is a kinetically controled process. This is due to the fact that dimer 2 is higher in energy that the first one. This indicates that the transition state is more stable in the is case than in the other. The energy difference is not great, with most components being the very similar. The main difference is in the Torsion of the second dimer
| Monohydrogenated 3 | monohydrogeneated 4 | ||||||
|---|---|---|---|---|---|---|---|
|
|
| output data from MM2 calculations | structure 3 | structure 4 |
|---|---|---|
| Stretch | 1.2067 | 1.1034 |
| Bend | 18.8637 | 14.5151 |
| Stretch-Bend | -0.7528 | -0.5464 |
| Torsion | 12.2396 | 12.5123 |
| Non-1,4 VDW | -1.5532 | -1.0470 |
| 1,4 VDW | 5.7649 | 4.4985 |
| Dipole/Dipole | 0.1632 | 0.1408 |
| Total Energy (Kcal/mol) | 35.9322 | 31.1767 |
By looking at the data for the monohydrogenated products it is obviouse that the second structure (4) is the more favoured one. Lookin at the different contributions to the energy it is easy to see where the change has come from. The difference is streching energies is very little, the second strucutre is slightly more ridgid. The main difference comes from the bend energies, where there is 4 Kcal/mol difference, so indicates that the second structure is far more ridgid, due to the fact that the double bond is on the five membered ring, the other 6 membered ring is keeped rigid by the bridge. In the other the double bond is on the six membred ring, so leaves the five membred ring less stable and places strain on the ring it is in. Both molecules have very similar torsion as the conformations are very similar, so where the double bond is, does not affect the structure much. The van der waal's terms vary, but when combinded they balance out between the two molecules.
Stereochemistry of Nucleophilic additions to a pyridinium ring (NAD+ analogue)
| structure 5 | structure 7 | ||||||
|---|---|---|---|---|---|---|---|
|
|
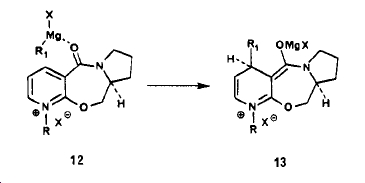
The reason for the absolute steriochemistry found in the product of reacting compound 5 with MeMgI is due to chelation of the magnesium ion by the carbonyl group in the transition state of the reaction. Given that in the structure the amide carbonyl is held up by the conformation then the Me group is delivered to the top face of the pyridine ring. When the Me is added a magnesium enolate also forms, The MgX then leaves to remove the positive charge for the ring N and release the counter ion, which then coordinates to the MgX cation. A way to get a better idea of the steriochemistry would be to model the transition state or intermidate where the MgX is bonded to the carbonyl to form the magnesium enolate. The diagram below shows the schem proposed by A.Schults etal[1].
Stereochemistry and Reactivity of an Intermediate in the Synthesis of taxol
| structure 10 | structure 11 | ||||||
|---|---|---|---|---|---|---|---|
|
|
Both molecules where drawn using the chem draw part of chemBio 3D. They where then both optimised using the MM2 program. In both cases the structures where not ideal, so the structures where alltered using chembio 3D, and then optimised. The main thing was getting the cyclohexane ring to form a chair structure, which ChemBio 3D does not seem to do on its own.
Hyperstable alkenes Hypersability of a cyclic alkene occours when the strain energy of the alkene ring is lower than the starin energy of the corisponding alkane ring. This is measured using olofine strain energy (OST). In most cyclic cases this is possitive, indicating the alkene is less stable than the alkane. In hyperstable cases the OST is negative, indicating it is more stable than expected. This is due to hydrogen interactions in the ring, they can be transanular or spacial interactions. The consequence of hyperstability is to render the alkene very unreactive to catalytic hydrogenation.[3]
This can be shown in this case. In the table below is the MM2 energies for the two structures and there hydrogenated counter parts
| structure 10 | structure 11 | |
|---|---|---|
| alkene | 48.9090 | 44.3159 |
| alkane | 72.5185 | 54.9140 |
These energies clearly show that both molecules are more stable as the alkene compared to the alkene, this shows that these structures are both hyperstable alkenes.
How one might induce room temperature hydrolysis of a peptide
Both structures where drawn on ChemBio 3D. For both structures an inital molecule was drawn and from that a copy was taken and the two different conformers where made from there. This took many attempts as the program genrally changed what I put in. the hardest part was getting the two rings to both be in a chair-chair arrangment. We use of the atom moving tool and reminimisation all four structures where obtained as chair-chair arrangments. The fact that the MM2 method regually produced structures with chair-twist chair conformations indicates that the energy barrier between equatoral and axial is not great and that the energy of the two are similar, which is why the program did not easily produce a chair-chair conformation
| Axial | Equatorial | ||||||
|---|---|---|---|---|---|---|---|
|
|
| Axial | Equatorial | ||||||
|---|---|---|---|---|---|---|---|
|
|
By looking at the structures it is easy to see why structure 14 does not react very readily, there is significant spacial speperation between the hydroxy and the carbonyl group, in one case as a strong H bond between the hydroxy group and carbonyl group.
The differecne in reactivity is explained by the fact that in each compound one of the conformations is more stable than the other. In structure 13, is is the equatoral structure that is more stable, and in this conformation the reaction can occour. In structure 14 the stable conformation is the axial one, which is not the reactive species. This indicates that under the reaction conditions used it is a kinetic reaction for 13 and thermodynamic for 14. This explains the dermatic difference in reactivity.
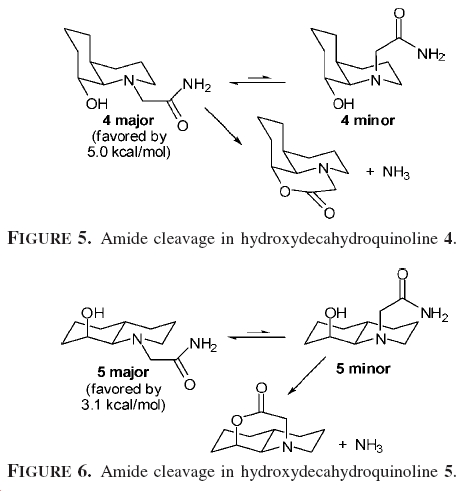
The paper by Nicolette M. Fernandes, Fabienne Fache, Mari Rosen, Phuong-Lan Nguyen, and David E. Hansen came up with a similar idea, there proposed idea for the differecne in reaction times is shown below
In the paper they ran MM2 calculations to try to work out the more stable forms of each isomer. Below I will compare the difference in energy values I obtained with the ones given in the paper.
| predicted | paper | |
|---|---|---|
| 13 | 7.7757 | 5.0 |
| 14 | 9.0489 | 3.1 |
while they where correct to say that the two axial products where more stable, But there must have been somthing wrong with there method as the energies are greater than there prediction. Also there value of 14 being favoured is relativily small. But the larger value I obtained explains the drematic difference in reaction half lives.
Modelling Using Semi-empirical Molecular OrbitalTheory
Regioselective Addition of Dichlorocarbene
| MM2 | HF/STO-3G | B3LYP/6-31G(d) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
| |||||||||
| c=c-c bond angle | |||||||||||
| 123.4553 | 123.6640 | 123.8261 | |||||||||
| H-C-Cl | |||||||||||
| 107.0784 | 109.3060 | 90.1495 |
Though the three different different structures look very similar, there are some significant differences in the angles produced within the molecule. There is some slight difference in the ring angles, which is due to the different methods used. The greater difference is in the angle between the H and Cl. this has a variation of 19 degress, which is significant, but it also indicates that the exact possition of the H and Cl have relativily little effect on the whole molecule.
The images below show the molecular orbitals HOMO-1 through to LUMO+2
| HOMO-1 | HOMO | LUMO | LUMO+1 | LUMO+2 |
|---|---|---|---|---|
 |
 |
 |
 |

|
When compared to the MO's predicted by Brian Halton, Roland Boeseb and Henry S. Rzepa[5]
frequencey data was calculated using the method and basis set given in the instructions: # b3lyp/6-31G(d) opt(maxcycle=30) freq Below is a table of the streching frequenceies which apply to the c-c double bonds and the Cl-C bond
| frequency (cm-1) | assigment |
|---|---|
| 1760.91 | exo double bond stretch |
| 1740.76 | endo double bond stretch |
| 772.634 | C-Cl bend |
monohydrogonated dichlorcarbene |
The structure of the monhydrogonated anti product was optimised in the same was as the dichlorocarbene. The final optimised structure is shown above.
Below are the IR frequencies for the CC double bond and the C-Cl bond.
| frequency (cm-1) | assigment |
|---|---|
| 1740.63 | endo double bond stretch |
| 757.679 | C-Cl bend |
Mini Project
introduction
I chose to look at the regioselectivity of the Baeyer-Villiger reaction in the production of beta-lactam antibiotics[6]. The reaction is intresting as it can give a range of products depending on what the R subsituent is.
In the paper they used a number of different R groups. In this project I will model and run calculation on the molecule where R=ipr. The reason for this is that with the other groups used, only one product was formed. The reaction initaly forms product 11, but the R group can migrate and change the product. ipr is an intermedate grup and so give close to a 50:50 mixture of products.
| product 10d | product 11d | ||||||
|---|---|---|---|---|---|---|---|
|
|
Both structures where constructed using ChemBio 3D and the structures optimised using MM2. This was then improved on by using Gaussian using DFT mpw1pw91 and basis set 6-31G(d,p). The calulatins took 1 hour 5 minutes for product 11d and 1 hour 14 minutes for product 12d. The structures shown above are the fully optimised structures.
NMR predictions
The optimised structure log files where down loaded form SCAN and then saved as gaussian input files. The text file was opened and the top edited as directed in the web page to give the NMR assignment. The method being used was the same and the solvent was chloroform, which is what was used in the original paper. Both structures where submitted to SCAN. Both calculations took about 7 minutes 30 seconds. The log files where then downloaded and opend in Gausse view and the predicted NMR found. For all the NMR's I chose the refrence TMS mPW1PW91/6-31G(d,p) CDCl3 GIAO as this refrence was calculated using the same method as I had used to calculate the NMR spectra.
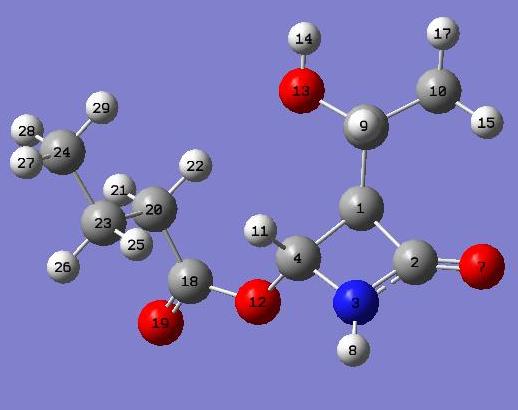
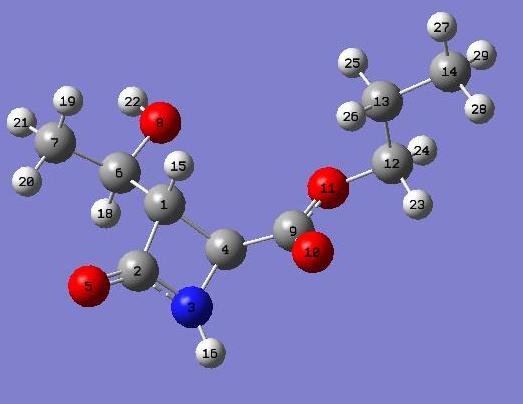
For both the carbon and hydrogen NMR the carbons and protons are lablled. Below is an image of each product with all atoms labled.
Carbon NMR
The peak list for both products can be found in the supporting material PDF, but no full spectra are provided, given the products are produced as a mixture and no method is explicity stated as to how the two products where seperated.
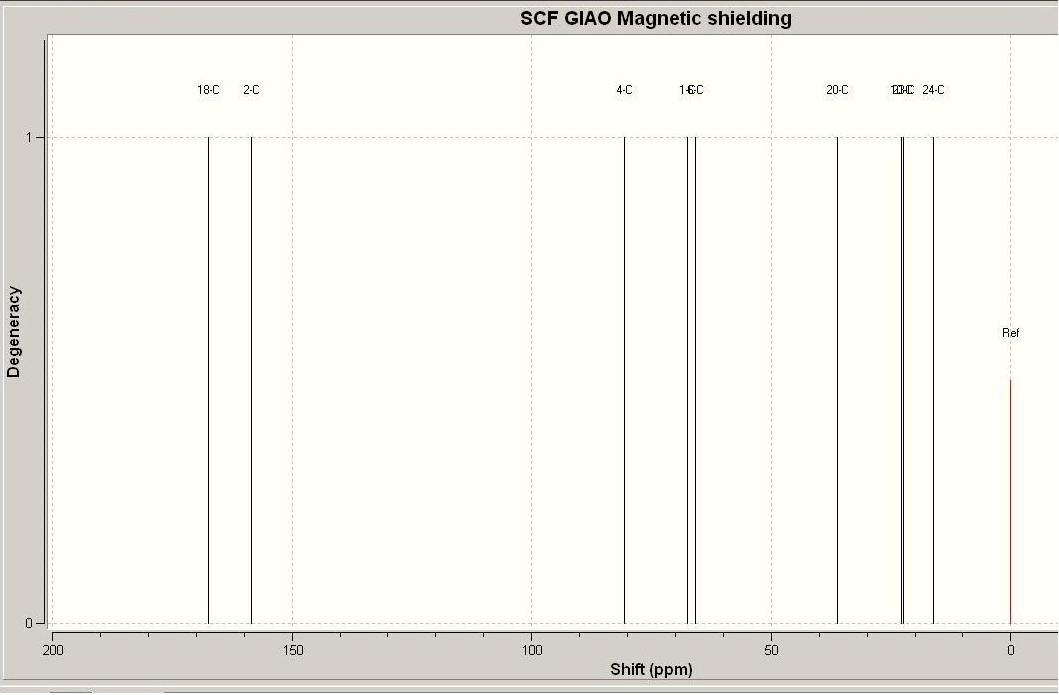
Product 11d NMR compared to predicted NMR
| Carbon | predicted NMR [7] | litriture NMR[8] |
|---|---|---|
| C1 | 67.5 | 65.3 |
| C2 | 158.5 | 167.2 |
| C3 | 80.63 | 75.9 |
| C4 | 80.6 | 75.9 |
| C6 | 65.8 | 63.9 |
| C10 | 22.8 | 21.3 |
| C18 | 167.4 | 177.6 |
| C20 | 36.8 | 24.0 |
| C23 | 22.4 | 18.9 |
| C24 | 16.2 | 18.8 |
The difference between the predicted and actual C NMR is significant in some cases, and in others not. The cases where the greatest difference occours is where the carbons are surrounded by several electron rich oxygen atoms. The fact that there is a difference (up to 10 ppm) is not supprising in these cases as the carbons would be heavily shielded, hence the high chemical shifts.
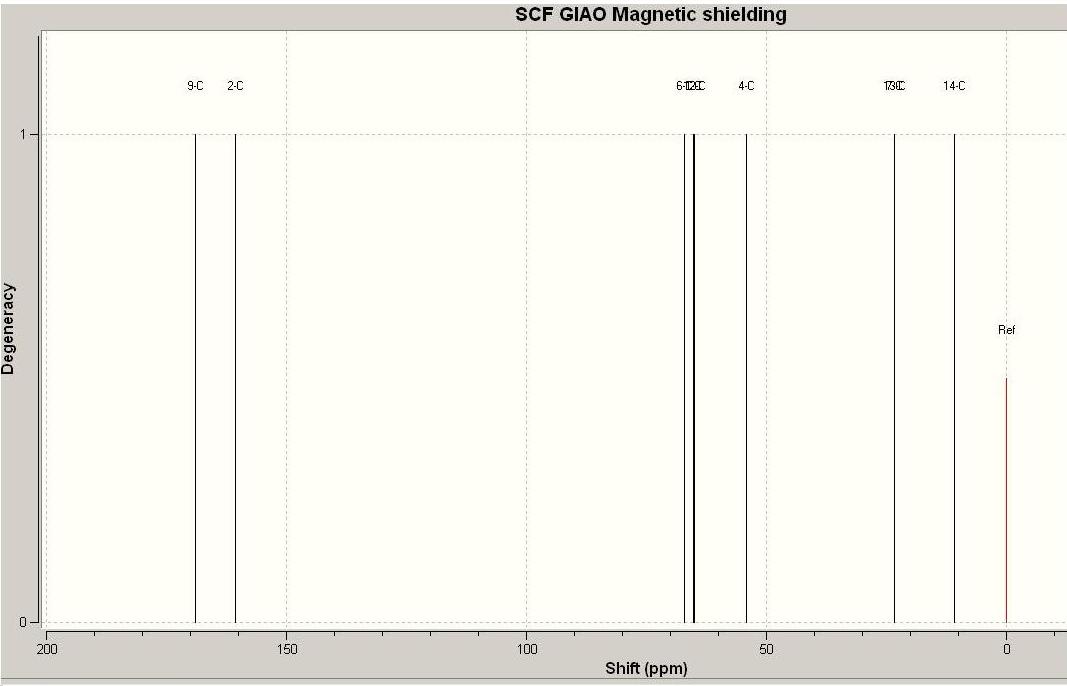
Produt 12d C NMR data compared to litriture
| Carbon | predicted NMR [9] | litriture NMR[8] |
|---|---|---|
| C1 | 65.2 | 64.5 |
| C2 | 160.6 | 168.3 |
| C4 | 54.0 | 50.0 |
| C6 | 65.2 | 64.0 |
| C7 | 23.2 | 21.8 |
| C9 | 169.0 | 170.8 |
| C12 | 65.0 | 69.7 |
| C13 | 23.4 | 21.9 |
| C14 | 11.0 | 21.3 |
Again there are errors associated with the data, but they are all within 10 ppm so are within the limits of the method used.
H NMR coupling constants calculated using janocchio
Product 11 H9 to H17,16,15 5.77 Hz H9 to H5 9.9 Hz H8 to H11 1.01 Hz H21,22 to 25, 26 177.7 and 62.5 Hz H 25,26 to H 25, 27 and 28 3.25 Hz
Product 12 H18 to H19, H21 and H20 5.8 Hz H18 to H15 to 9.88 Hz H16 to H17 0.13 Hz H23,H24 to H25,H26 11.1 and 5.4 Hz
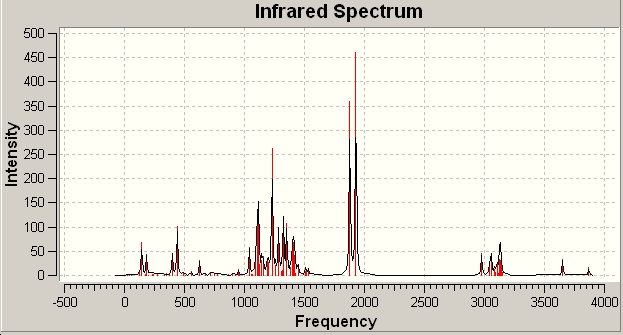
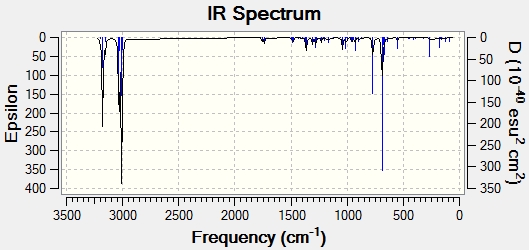
Predicted IR spectra
Product 11d predicted IR
The spectrum did produce a couple of negative vibration modes, so the structure may not be a fully optimised structure, but a transition state. The frequency analysis did produce the destinctive C=O stretches at 1877.7 and 1929 cm-1 for the ester group and the ring carbonyl
Product 12d predicted IR
As with the other product the spectra produced had some negative vibrational modes, so indicating the structure is a transition state, which given the molecule is very flexible, that is not supprising that it did not mimimise given the method and basis set used. This again gave two different C=O stretches at 1850.8 and 1922.7 cm-1 for the ester group and the ring carbonyl
Conclusion
While the method used worked well, the fact that the structure was not fully optimised means that the NMR and IR data may be unreliable. But when compared to litriture values the fit is relatiivily good. Both compounds are very similar and so would be hard to seperate, but one seperated use of H and C NMR with IR would give positive identification of the two products.
refrences
- ↑ : Regio- and stereoselective control in the addition of Grignard reagents to the pyridine ring system DOI:10.1021/jo00356a016
- ↑ : Regio- and stereoselective control in the addition of Grignard reagents to the pyridine ring system DOI:10.1021/jo00356a016
- ↑ Pentacyclo[8.2.1.12"~.14'7.1s'H]hexadeca-l,7-diene and Its 4,5,10,11-Tetramethyl Derivative, Two Highly Hyperstable Slightly Pyramidalized Dienes:DOI:10.1016/S0040-4020(97)00595-4
- ↑ Rapid Cleavage of Unactivated, Unstrained Amide Bonds at Neutral pH: DOI:10.1021/jo800706y
- ↑ A molecular orbital and crystallographic study of the structure and -facial regioselectivity of 9-chloro-1,4,5,8-tetrahydro-4a,8a-methanonaphthalene: DOI:10.1039/P29920000447
- ↑ 6.0 6.1 Regioselective Baeyer−Villiger Oxidation in 4-Carbonyl-2-azetidinone Series: A Revisited Route toward Carbapenem Precursor: DOI:10.1021/jo030377y
- ↑ product 11d NMR data: http://hdl.handle.net/10042/to-1225
- ↑ 8.0 8.1 Regioselective Baeyer−Villiger Oxidation in 4-Carbonyl-2-azetidinone Series : A Revisited Route toward Carbapenem Precursor supporting information: http://hdl.handle.net/10042/to-1225
- ↑ product 12d NMR data: http://hdl.handle.net/10042/to-1225