Rep:Module1:rp1207
Endo and Exo Cyclopentadiene Dimer Comparison

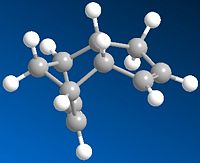
Cyclopentadiene dimerises via a [2,4] Diels Alder Reaction to produce molecule 2,the endo dimer specifically despite the greater stability of molecule 1, the exo dimer.
. The two dimers were modelled using ChemBio 3D and their energies minimalised usign the MM2 their relative energies could then be compared. As seen in the table below molecule 2, the exo dimer has the lowest total energy of the two, however is not the major product of the reaction, therefore it can be assumed that the reaction proceeds by kinetic control, whereby the reaction proceeds not in favour of the lower energy thermodynamic product but via a lower energy transition state to produce the less stable endo dimer.
This is unsuprising for a cycloaddition reaction as they usually proceed via stereoelectronic control and reactions proceed according to molecular orbital overlap.
The table below not only elliterates the stability of molecule 1 in comparison to molecule 2 but also isolates the most destablising factor in Molecule two which can be seen to be the torsion angle. This, as intuition would suggest, shows that it is bond strain, particually as seen for the carobon bridge that is largely responsible for the difference in energies between the two products.
| Interaction | Molecule 1 | Molecule 2 | |
|---|---|---|---|
| Stretch / K cal Mol-1 | 1.2817 | 1.2549 | |
| Bend / K cal Mol-1 | 20.5695 | 20.8638 | |
| Stretch-Bend / K cal Mol-1 | -0.8364 | -0.8375 | |
| Torsion / K cal Mol-1 | 7.6672 | 9.5055 | |
| Non 1,4- VDV / K cal Mol-1 | -1.4354 | -1.5314 | |
| 1,4-VDV / K cal Mol-1 | 4.2354 | 4.3013 | |
| Dipole Dipole/ K cal Mol-1 | 0.3778 | 0.4461 | |
| Total Energy / K cal Mol-1 | 31.8795 | 34.0027 |
Cyclopentadiene Dimer Hydrodgenation


As can be seen from the table below the most stable of the Cyclopentdiene Hydrogenated products is Molecule 4 which has a total energy ~31.2 KcalMol-1 which is low relative to the total energy of molecule 3 which ~35.7KcanMol-1, therefore from this investigation it can be concluded that Molecule 4 is the thermodynamic product of hydrogenation. The kinetic product cannot be predicted from these results, further investigation would be required.
The main energy differences between invividual interaction energies are those of the bending energies which are more destablilising for Molecule 3 and can be seen as the main destabilising force responsible for the higher total energy fo the molecule.
Interestingly Molecule 3 shows lower torsion energy than Molecule 4 which suggests that the bond angles are less strained in this product, however this energy difference is clearly counterbalanced by the general higher energies in most other indervidual interactions.
| Interaction | Molecule 3 | Molecule 4 | |
|---|---|---|---|
| Stretch / K cal Mol-1 | 1.2659 | 1.0990 | |
| Bend / K cal Mol-1 | 19.8063 | 14.5133 | |
| Stretch-Bend / K cal Mol-1 | -0.8276 | -0.5477 | |
| Torsion / K cal Mol-1 | 10.8698 | 12.5076 | |
| Non 1,4- VDV / K cal Mol-1 | -1.2207 | -1.0512 | |
| 1,4-VDV / K cal Mol-1 | 5.6394 | 4.5070 | |
| Dipole Dipole/ K cal Mol-1 | 0.1621 | 0.1407 | |
| Total Energy / K cal Mol-1 | 35.6953 | 31.1687 |
Stereochemistry of Nucleophilic Additions to a Pyridinium Ring
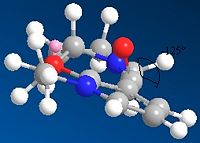
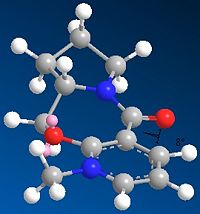
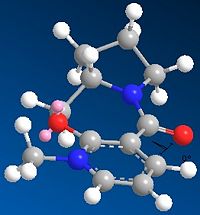
Molecule 5
In this part of the investigation the relative energy values of Prolinol were studied in relation to the dihedral angle between the carbonyl group and the 4-position on the pyridine ring. This was achieved by creating the reactant molecule in ChemBio3D and minimising the energy using the MM2 force field, this was repeated for several different geometries until the lowest was found. The geometry of the reactant can then be compared to the geometry of the product and a reasonable mechanism for the reaction suggested.
As can be seen from the table below the most stable form of the molecule is that with a dihedral angle of 9°• is the most thermodynamically favoured of those investigated.
| Dihedral Angle | 125 | 8 | 9 | |
| Stretch: kcal/mol | 3.846 | 2.061 | 2.0507 | |
| Bend: kcal/mol | 112.1606 | 14.3051 | 14.3267 | |
| Stretch-Bend: kcal/mol | -0.297 | 0.1317 | 0.133 | |
| Torsion: kcal/mol | 28.5468 | 5.0381 | 5.045 | |
| Non-1,4 VDW: kcal/mol | 0.644 | -0.4472 | -0.4753 | |
| 1,4 VDW: kcal/mol | 28.4111 | 16.5717 | 16.5327 | |
| Charge/Dipole: kcal/mol | -8.5087 | 9.5293 | 9.5463 | |
| Dipole/Dipole: kcal/mol | -3.4717 | -4.0318 | -4.0222 | |
| Total Energy: kcal/mol | 161.3311 | 43.1579 | 43.1369 | |
As can be seen from the image the 125°• bond angle results in large 1,4-steric strain, this can be seen as the 1,4-Van Der Waals energy for the 125 molecule ≈29Kcal mol-1 whereas for the lower dihedral angles the 1,4-Van Der Waals energy≈17 Kcal mol-1
The favouring of the 9° distortion of the carbonyl ring within the same face as the puckering of the seven membered ring suggests the reason why the absolute stereochemistry of the reaction occurs.
The reaction proceeds via coordination of the grignard reagent with the Prolinol ring; the Co-ordinating methyl group with the 4 position of the Pyridine ring and the coordination of the lewis acid, Mg , to the carboxyl group. Due to this coordination the methyl group will bind to the ring along the same face as the carboxyl group. Resulting in the indicated product.
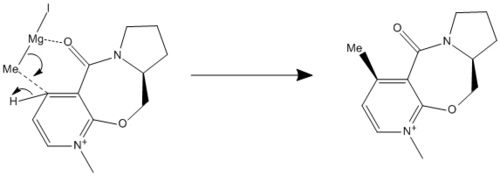
As the most favoured ground state for the prolinol molecule is that with the carboxy group pointing out of the plane in the same direction as the puckering of the seven membered ring, therefore the grignard reagent coordinates on this site and the methyl group will attach to the 4 position of the Pyridine ring on this face.
Molecule 7

As can be seen from the table below the optimum dihedral angle for the carbonyl group in the 7 membered ring is 22, as can be seen from the image this relates to the carbonyl stretching up onto the opposite face to the adjacent methyl group. This provides a good explanation for the geometry of the product as both the carbonyl group and the NHPhenyl group coordinating with the pyridinium ring have large electron deinsity which will repel each other, therefore the NHPhenyl group will coordinate on the oppisite face to the carbonyl, therefore based on the proposed geometry of the start material it will eb expected to coordinate on the same face as the mathyl group, this prediced relationship is in agreement with the structure of molecule 8.
| Dihedral Angle | 22 | -19 | |
| Stretch: kcal/mol | 3.9606 | 3.9447 | |
| Bend: kcal/mol | 11.6941 | 11.7486 | |
| Stretch-Bend: kcal/mol | 0.4072 | 0.4068 | |
| Torsion: kcal/mol | 9.6889 | 9.6603 | |
| Non-1,4 VDW: kcal/mol | 4.1599 | 4.16012 | |
| 1,4 VDW: kcal/mol | 29.2976 | 29.3483 | |
| Charge/Dipole: kcal/mol | 9.0594 | 9.0160 | |
| Dipole/Dipole: kcal/mol | -4.8819 | -4.8861 | |
| Total Energy: kcal/mol | 63.3858 | 63.3988 | |
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
This Taxol intermediates shown may occur in two possible carboxyl isomers. These have been modelled using ChemBio 3D and their energies compared using MM2 in order to determine the most stable isomer.
The most stable conformation of each form was found by rearranging the bond in each molecule and minimising the energy using the MM2 technique until the lowest total energy value was found.
As can e seen from the table below molecule 10 has a significantly lower total energy and is therefore the nmost stable conformatino of this product. The main factors contributing to this increased stability is a lower beding energy-bending energy for Molecule 10 ~10.8 Kcal compared to Molecule 9 which has a bending energy ~20.5, a reduction of almost half! The torsional strain of molecule 9 is seen to be significantly higher than that of molecule 10, this is discussed in more detail below.There is also a significant difference in the 1,4 VDV energies of the molecuel which contributes to the overall stability of molecule 10.
As can be seen in the produced images, one of the main differences between the two molecules is that molecule 9, in order to produce a carbonyl which faces 'upwards' in the same face as the bridging group must adopt a relatively unstable twist boat conformation in the 6 memebred rign whereas the geometry of the carbonyl group in molecule 10 allows the six memebered rign to form the less sterically and torsionally strained chair conformation. This is reflected in the relative energies discussed above and seen in the table below.
| Molecule Number | 9 | 10 | |
| Stretch: kcal/mol | 3.4777 | 2.5822 | |
| Bend: kcal/mol | 20.5371 | 10.7855 | |
| Stretch-Bend: kcal/mol | 0.4913 | 0.3221 | |
| Torsion: kcal/mol | 23.2927 | 19.7458 | |
| Non-1,4 VDW: kcal/mol | 0.3252 | -1.4646 | |
| 1,4 VDW: kcal/mol | 15.7820 | 12.5345 | |
| Dipole/Dipole: kcal/mol | 0.0667 | -0.1819 | |
| Total Energy: kcal/mol | 63.9727 | 44.3237 | |
The alkene functional groups in these molecules react slowly upon electrophilic attack. This is because they nmay be described as hyperstable alkenes, as the kinetic stabliity of bridgehead alkenes results in them resisting sp2 to sp3 hybridisation. This is founded on the concept of olefin strain (OS) which can be calculated as; Strain energy of most stable confomer of hydrocarbon-Strain energy of olefin Systems which result in negative OS values, with greater strain in conformer than in the origional olefin, are known as hyperstable alkenes.
Modelling Using Semi-empirical Molecular Orbital Theory
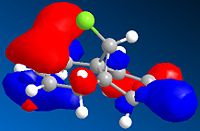
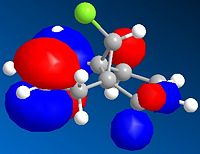
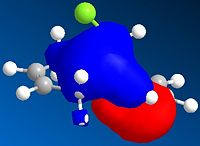
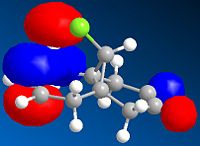
| HOMO-1 | HOMO | LUMO | LUMO +1 | LUMO +2 | |
 |
 |
 |
 |
||
 |
 |
 |
 |
||
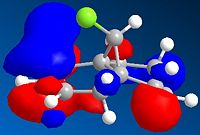
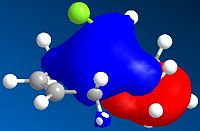
It can be seen from the above images that the Alkene that lies under the C-Cl bond will be the most suceptible to electrophilic attack. This is because there is a large amount of electron density dispersed about the bond. This could suggest that as thsi double bond, which as a cloud of electron density between itself and the C-Cl bond suggesting it is donatign electron density to the electronegative Cl atom, is itself drawing electron density from the adjacent C=C bond.
As can be seen from the above table the main difference between the two molecules (apart from the obious lack one on C=C bond) is the increase in wavenumber for the C-Cl bond, this suggests that the C-Cl bond is stronger for the monoalkene than the dialkene.
This can be explained using Molecular Orbital theory.
As can be seen from the above images the HOMO for the 2-alkene product the electron density lies above the plane of the C-C between the C-C bodn and the Cl atom, this suggests that this alkene group donates electro density into the C-Cl bond thus weakening it and being responsible for the weaker C-Cl bond in the diene.
This does not occur with the single alkene as this is not in the correct orientation to donate into the C-Cl antibonding orbital.
Mini Project
Within this project I whave investigated the reduction of a 4-Piperidonone, the mechanism for this resuction using L-Selecride3, discussion of how the resulting isomers may be identified using spectroscopic techniques and comparison of computer estimated spectroscopic values with those found in literature.
The reaction for the Reduction of 3-Isopropenyl-1-(toulene-4-sulphonyl)-piperidin-4-one is shown below;
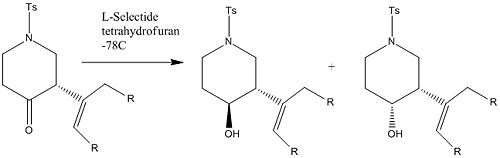
The Products
The two products are shown pictorally below;
The energies of the optical RS and RR are very similar with only slight differences, this is due to the large similarities in structure. A table summarising the total energies of each molecule and the components contributing to this total is shown below;
| Isomer | 3R 4S | 3R 4R | |
| Stretch: kcal/mol | 1.1618 | 1.1638 | |
| Bend: kcal/mol | 2.6340 | 2.9758 | |
| Stretch-Bend: kcal/mol | 0.2118 | 0.2282 | |
| Torsion: kcal/mol | -3.2718 | -2.4875 | |
| Non-1,4 VDW: kcal/mol | -7.4721 | -6.6166 | |
| 1,4 VDW: kcal/mol | 15.8432 | 15.8149 | |
| Dipole/Dipole: kcal/mol | 1.4837 | 1.2159 | |
| Total Energy: kcal/mol | 10.5907 | 12.2945 |
As can be seen from the above table the 3R4S product is the thermodynamic product of the reduction, this is large due to some relieving of the non-1,4-Van Der Vaals strain which occurs between teh Hydroxyl and Alkyl groups.
However literature suggests that use of a iBu2AlOiPr reducing agent in toluene the 3R4R product may be the major product by a ratio of 99:1. This suggests that under these reaction conditions the kinetic product is formed.
This is supported by the chosen reducing agent L-Selectride which, as shown below, has large bulky groups which will prevent it from reducing the carbonyl group on the same face as the neighbourign R group.
Dipole Moment
| 3R 4S Dipole Moment/Debye | 3R 4R Dipole Moment/Debye | |
| 4.3648 |
Due to the large length of time to compute the Dipole moment and computer errors only the dipole moment for the RS Isomer has been calculated however it would be expected that the RR isomer would have a greater dipole moment as the alkene and -OH groups would be pulling electron density in a similar direction resulting in a greater overall dipole moment for the molecule as a whole.
This would be a good way to distinguish between the two isomers as the only difference between the two molecules is the geometry of the -OH group, one in the same directino as an alkene and one in an opposite face. Also, literature values would not nessisarily be nessisary to draw a conclution as the two molecules in relation to each other can be used to distinguish between them (if both were present).
Optical Rotation would also be a good method to distingush between the two products at they each have two stereocentres with one differing between them, a literature value for these figures is not available and the programme used to predict these values computationally is yet to result in a successful output within the set timescale.
3R 4S
'13CNMR'
| Carbon | Literature Chemical Shift/ppm | Calculated Chemical Shift/ppm | % Error | |
| C1 | 21.5 | 23.3 | 8.4 | |
| C2 | 22.8 | 27.7 | 21.5 | |
| C3 | 31.1 | 35.9 | 15.4 | |
| C4 | 40.5 | 44.8 | 10.6 | |
| C5 | 43.5 | 47.3 | 8.7 | |
| C6 | 46.7 | 54.9 | 17.6 | |
| C7 | 63.0 | 65.6 | 4.1 | |
| C8 | 112.2 | 113.7 | 1.3 | |
| C9 | 127.6 | 117.7 | 8.4 | |
| C10 | 129.6 | 118.4 | 9.5 | |
| C11 | 133.4 | 127.1 | 5.0 | |
| C12 | 143.4 | 131.2 | 9.3 | |
| C13 | 143.8 | 142.6 | 0.8 |
4
The calculated NMR for this product shows reasonable agreement with literature values with a maximum-one off-error of 21% and all others lying below 15% error and an average overall error of 7.9%. The suggests that the molecule has been modelled within a reasonable degree of accuracy and the large errors may have occured due to the computer programme not taking into account attached hetero atoms for example C-1st row TM can have an error ~3ppm. Errors may also have occured if there are isolated areas within the moelcuel where the geometry has not been fully optimised due to the rough approximation of MM2.
'IR'
| Assigned Bond | Literature Wavenumber/cm-1 | Calculated Wavenumber/cm-1 | Error % | |
| O-H | 3519 | 3757.34 | 6.8 | |
| 2920 | 2999.80 | 2.7 | ||
| 2904 | 2945.54 | 1.4 | ||
| Symmetic Aromatic C-C Stretch | 1651 | 1655.27 | ||
| Aromatic C-C Stretch | 1597 | 1537 | 3.9 | |
| C-H Wag | 1447 | 1447.0 | 0 | |
| C-H wag | 1323 | 1316.96 | 0.5 | |
| 1304 | 1306.68 | 0.2 | ||
| C-O Stretch | 1157 | 1156.34 | 0.1 |
The IR stretches also show particually good areement with the literature values suggesting that this molecule has been optimised within an excellent accuract and that IR stretches may by predicted within a reasonable accuracy. SOme of the error occured in both the NMR and the IR may also be due to errors in experimental data. As the values for the RR isomer can be seen to have similar accuracy at a glance, a percentage error value has not been calculated for these values and they have been assumed to be go a good accuracy.
3R 4R
| Carbon | Literature Chemical Shift/ppm | Calculated Chemical Shift/ppm | |
| C1 | 20.6 | 27.6 | |
| C2 | 21.5 | 35.4 | |
| C3 | 32.5 | 41.8 | |
| C4 | 45.0 | 43.8 | |
| C5 | 48.8 | 49.2 | |
| C6 | 51.6 | - | |
| C7 | 69.4 | 72.1 | |
| C8 | 114.7 | 113.1 | |
| C9 | 127.6 | 117.7 | |
| C10 | 129.7 | 118.5 | |
| C11 | 133.6 | 129.4 | |
| C12 | 142.4 | 130.1 | |
| C13 | 143.6 | 144.6 |
5 IR
| Assigned Bond | Literature Wavenumber/cm-1 | Calculated Wavenumber/cm-1 | |
| Asymmetric C-H Stretch | 3391 | 3230.49 | |
| C-H Stretch | 2920 | 3030.83 | |
| C-H Stretch | 2904 | 3006.53 | |
| C-C Aromatic Stretch | 1647 | 1655.33 | |
| C-C Aromatic Stretch | 1597 | 1537.26 | |
| C-H Wag | 1458 | 1427.04 | |
| C-H Wag | 1334 | 1325.65 | |
| C-H Bend | 1304 | 1305.73 | |
| 1157 | 1166.28 |
Reference List
1.http://pubs.acs.org/doi/pdf/10.1021/cr00023a007 Maier, W. F.; Schleyer, P.v.R. J. Am. Chem. Soc. 1981,103,1891. 2.McEwen, A. B.; Schleyer, P.v.R. J. Am. Chem. SOC1. 986,108,3951.
3. Steven W. Elmore1 and Leo A. Paquette,*
4. DOI:10042/to-2963 5. DOI:10042/to-2964