Rep:Module1:jzhao
The basic techniques of molecular mechanics and semi-empirical molecular orbital methods for structural and spectroscopic evaluations
Aim of this section:
- To use molecular mechanics to predict the geometry and regioselectivity of three reactions.
- To use semi-empirical and DFT molecular orbital methods to investigate structure and spectroscopy.
Modelling using Molecular Mechanics
The Hydrogenation of Cyclopentadiene Dimer
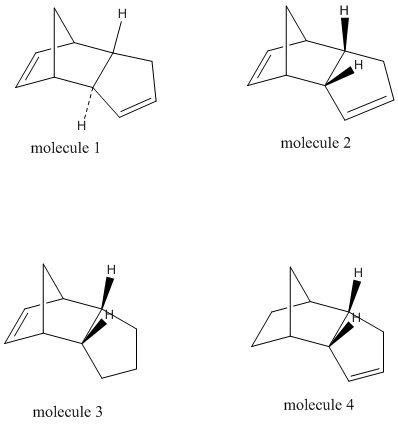
The optimisation of the geometries of molecule 1 and 2 using MM2 force option (Calculations/MM2/Minimise energy) gives the total energy to be 31.8832 kcal/mol for molecule 1 and 34.0028 kcal/mol for molecule 2, which means that dimer 1 is thermodynamically more stable (less strained) than dimer 2. However, cyclopentadiene dimerises to produce specifically the endo dimer 2 rather than the exo dimer 1 according to the literature1[1]. This indicates that the cyclodimerisation of cyclopentadiene reaction is kinetically controlled, leading to the product with lower energy trasition state.
Jmol models of molecules 1 and 2
The same optimisation of the geometries procedures was carried out for hydrogenation products, 3 and 4. The total energy is 35.9290 kcal/mol for molecule 3 and 31.1539 kcal/mol for molecule 4. As the total energy of molecule 4 is lower than molecule 3, the hydrogenation to form product 4 is thermodynaically more favourabe. If 4 is the major product, then the reaction is thermodynamically controlled, otherwise it is kinetically controlled. The relative contributions to total energy from the stretching (str), bending (bnd),torsion (tor), van der Waals (vdw) and hydrogen bonding (H-Bond) energy terms are concluded in the table below:
| Molecule 3 | Molecule 4 | |
|---|---|---|
| Stretching | 1.2247 | 1.0973 |
| Bending | 18.8939 | 14.5063 |
| Torsion | 12.1952 | 12.4976 |
| van der Waals | 5.7419 | 4.5127 |
| Dipole/Dipole | 0.3778 | 0.4465 |
| Hyrdrogen bonding | 0 | 0 |
The lagest contributions to the total energy is from bending and torsion. There is a lager difference in bending energy between the two molecules, which gives rise to the large difference in total energy.
Jmol models of molecules 3 and 4
Stereochemistry of Nucleophilic additions to a pyridinium ring (NAD+ analogue)
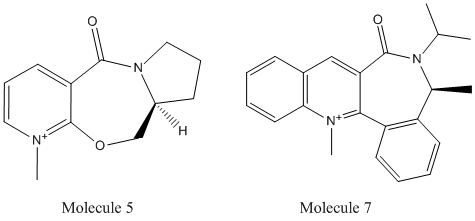
The models of the two pyridinium reactants 5 and 7 were constructed in ChemBio3D and minimised using MMFF94 force option (Calculations/MM2/Minimise energy). The MeMgI should not be included in constructing the model, as the MM2 calculation can not optimise the geometry for Mg atom. By varing the geometry of the two reactants, several geometries with slight differences investigated and optimised using the MMFF94 force option. The relationship between total energies and carbonyl group's orientations with respect to the aromatic ring (Structure/Measurements/Generate All Dihedral Angels)were concluded in the following table:
| Molecule 5 | Molecule 7 | ||
|---|---|---|---|
| Dihedral Angels | Total Energy (kcal/mol) | Dihedral Angels | Total Energy (kcal/mol) |
| 36o | 59.1431 | 40o | 113.091 |
| 27o | 58.9706 | 39o | 115.419 |
| 6o | 57.5653 | 35o | 99.6866 |
| 29o | 130.58 |
Many other geometries are possible (not listed in the table)
Jmol models of molecules 5 and 7
The reaction of 7 to form 8 can be explained by the geometry study. The nucleophilic reagent PhNH2 is relatively bulky. It attacks 7 in a way to avoid interacting with the C=O group (steric hindrance), which gives the product 8 with NHPh group pointing out of the plane. The opposite mechanism occurs for the MeMgI reagent, as the O atom on C=O can coordinate the Mg atom.
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
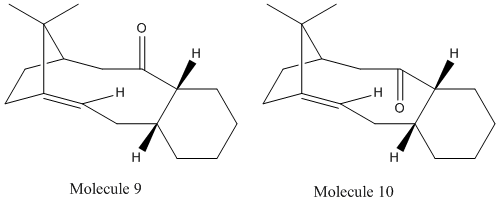
Jmol models of molecules 9 and 10
The optimisation of the geometries of isomer 9 and 10 using MM2 force option (Calculations/MM2/Minimise energy) gives the total energy to be 48.1642 kcal/mol for isomer 9 and 50.1079 kcal/mol for isomer 10. Isomer 9 is lower in energy, thus it is more stable. The functionalisation of the C=C double bond is abnormally slow. This is because molecule 9 and 10 are hyperstable alkenes.
Modelling Using Semi-empirical Molecular Orbital Theory
Regioselective Addition of Dichlorocarbene

1 Part 1: Orbital control of reactivity: reaction of compound 12 with electrophilic reagents The geometry of compound 12 was optimised firstly using MM2 force option (total energy: 17.8998 kcal/mol) and then using MOPAC interface (Calculations/MOPAC Interface/Minimise energy; select Method=PM6 from the Job & Theory pane; tick Molecular Surfaces from the Properties page). The molecular orbitals were viewed by selecting Surfaces/Choose Surface/Molecular orbital.
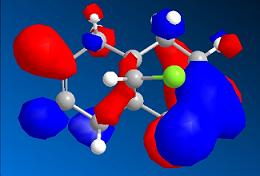
The HOMO of compound 12 shows differnet density on two C=C double bond, which indicates the two double bonds are chemically inequivalent. Therefore, this method can discriminate between the two alkene bonds. The HOMO shows a larger electron density on the alkene bond cis to the Cl-C bond, thus this alkene bond is more nucleophilic and more readily to undergo electrophilic addition. This is consistant with the literature 2[2].
.
2 Part 2: Calculation of the influence of the Cl-C bond on the vibrational frequencies B3LYP/6-31G(d,p)Gaussian geometry optimisation and frequency calculation were carried out with the MM2 optimised geometries of compound 12 and its dihydro derivative 13 using SCAN. The output FCHK files of dialkene 12 and monoalkene 13 obtained from the SCAN were loaded up into GaussView, and the vibrational modes of Cl-C stretching and C=C stretching were viewd and tabulated:
| Dialkene 12 | Monoalkene 13 | ||
|---|---|---|---|
| C-Cl stretching (cm-1) | 770.902, 901.522, 930.065, 952.343 | C-Cl stretching (cm-1) | 774.937, 908.673, 925.811 |
| C=C stretching (cis to Cl) (cm-1) | 1757.36 | C=C stretching (cis to Cl) (cm-1) | 1758.05 |
| C=C stretching (anti to Cl) (cm-1) | 1737.11 |
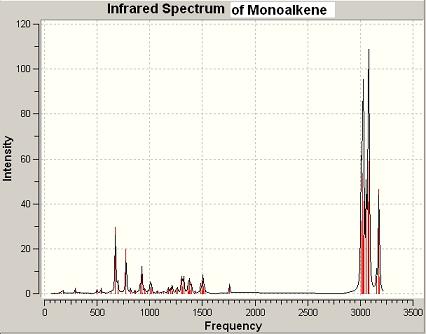
The C=C (cis to Cl )stretchings of dialkene 12 and dihyro-derivative 13 are very close in frequency, while the C=C (anti to Cl) stretching has lower frequency than the C=C (cis to Cl) stretching. This helps to support the relative reactivity of the two C=C double bonds in dialkene. The C=C (cis to Cl) has a larger electron density, which makes it a stronger bond than C=C (anti to Cl). Therefore, its stretching frequency is higher.
Structure based Mini project using DFT-based Molecular orbital methods
The objectives of the mini project
- to choose a reaction to study: the papers for the reaction must include experimental 13C for the ismeric products.
- to analyse the product using computational techniques to discuss:
- How to differentiate spectroscopically between the isomeric products?
- Calculate the predicted 13C spectra for the isomers.
- Compare the predicted data to the experimental data in literature.
Stereocontrolled Synthesis of (-)Cubebol
Introduction
Cubebol is a natural sesquiterpene alcohol first found in cubeb oil. The taste of cubebol is cooling and refreshing. It was patented as a cooling agent by an international flavorcompany, Firmenich in 2001. It is widely used in products ranging from chewing gum to toothpaste and drinks. The synthesis of cubel from (-)menthone is stereocontrolled process, involving several reaction intermediates2[3]. In this mini project, computational techniques were applied predict the NMR and IR spectra of cubebol. Since the syntesis of is fully stereocontrolled leading to the only product (-)Cubebol, we only need to predict the 13C NMR and IR spectra and compare them to literature data. If the predicted values are consistant with the literature values, it confirms that the product obtained to be (-)Cubebol.
Investigating conformational possibilities
Several conformational possibilities were investigated by varing the position of the atoms. The comformation with lowest energy, which has one 6-membered ring to be in chair comformation, was used for the following computational analysis.
Predicting the 13C NMR Spectrum
1 Geometry optimisation: The input file was created in ChemBio3D (Calculations/Gaussian Interface/Create Input File; Select Job Type/Minimise; Method DFT=mpw1pw91 ). The input file was modified before sending to SCAN for geometry optimisation.
2 NMR Chemical Shift calculation using scan: A new input file was created with the optimised geometry and modified before sending to SCAN for NMR chemical shift calcuation.
3 Analyzing the NMR Chemical Shift calculation: The fchk file from the SCAN was loaded up in GaussView and the NMR spectrum was viewed. The chemical shift of different carbons were tabulated and compared to the literature values.
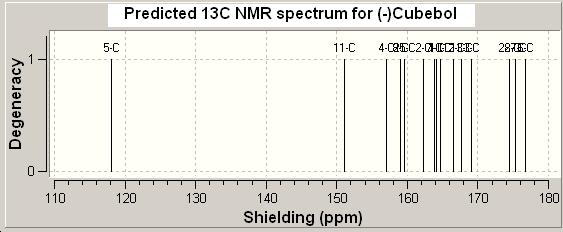
| Computational Analysis | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cheical shift | 176.8 | 175.3 | 174.6 | 169.0 | 167.6 | 166.5 | 164.6 | 164.3 | 163.8 | 162.3 | 159.5 | 159.2 | 157.0 | 151.7 | 118.2 |
| 195ppm - Cheical shift | 18.2 | 21.5 | 20.4 | 26.0 | 27.4 | 28.5 | 30.4 | 30.7 | 31.2 | 32.7 | 35.5 | 35.8 | 18.0 | 43.3 | 76.8 |
| Corresponding Carbon | C15 | C13 | C14 | C6 | C8 | C11 | C10 | C2 | C9 | C1 | C12 | C3 | C5 | C7 | C4 |
| Experimental (literature) | |||||||||||||||
| Cheical shift | 80.3 | 44.1 | 39.0 | 36.3 | 33.6 | 33.4 | 31.7 | 30.8 | 29.5 | 27.9 | 26.4 | 22.6 | 20.1 | 19.6 | 18.7 |
| Corresponding Carbon | C7 | C4 | C5 | C3 | C12 | C1 | C9 | C10 | C2 | C11 | C8 | C6 | C14 or C13 | C13 or C14 | C15 |
Large difference between the computational values and experimental values were observed. This is probably due to two factors: 1) the computational method is sensitive to the conformation of the molecule; 2) the different solvent systems used for 13C NMR spectrum. For the computational analysis, it was assumed that chloroform was used for 13C NMR. However, a different solvent might be used for the experimental spectroscopy (not mentioned in the literature). It was also found that 195ppm - chemical shift (obtained using computational analysis) was very close to the chemical shifts of the experimental values, which supports the idea that the chemical shift difference is due to the different solvent systems used.
Predicting the IR spectrum
Procedure: The input file was created and modified before sending to SCAN. Analyzing the Vibrational Spectrum: The the fchk file from the SCAN was loaded up in GaussView and the vibrational modes were viewed. The wavenumbers of different vibrational modes were tabulated and compared to the literature values.

| Computational Analysis | Experimental (literature) | ||
|---|---|---|---|
| Wavenumber (cm-1) | Vibrational mode | Wavenumber (cm-1) | Vibrational mode |
| 3492 (before correlation: 3796) | O-H stretching | 3350 (br) | O-H stretching |
| 3152-2980 | C-H stretching | 2951(m), 2860(m) | C-H stretching |
| 1528-909 | C-H bending | 1490(w), 1142(m), 910(w) |
Conclusion
The predicted 13C NMR and IR data using computation analysis thechniques do not match the experimental data perfectly. However, it provides useful additional information to check if the product obtained is the desired one, or to deduce a mechanism for a reaction. For the mini project - analysis of (-)Cubebol, the predicted 13C NMR and IR generally mathces the experimental data. The NMR shows large difference before correlation, probably due to the different solvent system used for the spectroscopy.
References
- ↑ J. Sauer, R. Sustmann, Angew. Chem., Int. Ed. Engl., 1980 , 19, 779 - 807. DOI:10.1002/anie.198007791
- ↑ B. Halton, R. Boese and H. S. Rzepa., J. Chem. Soc., Perkin Trans 2, 1992 , 447. DOI:10.1039/P29920000447
- ↑ D. M. Hodgson, S. Salik and D. J. Fox., J. Org. Chem. 2009