Rep:Mod:zyw0917
Computational Chemistry Lab Module 2
Yawen Zhang
In this inorganic chemistry lab, gaussview will be used to bulid up the molecules and the computational studies give the structure and bonding information of complexes. In recent studies, IR and Raman spectra, NMR spectra and dipole moments can also be analysed.
BH3 Introduction Module
creating a molecule
To build up a BH3 molecule, B atom was selected first and the trignoal planar orientation was then selected. A regular BH3 molecule with three equal B-H bonds would show.
Using the controls pallette, the bond distances, angles and torsion angles of molecule can be altered. The BH3 molecule shown below has been altered on B-H bond distance into 1.5 angstrom.
optimising a molecule
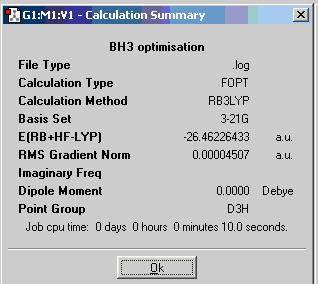
To optimise the BH3 molecule, B3LYP/3-21G/OPT method of Gaussian Calculation is used. The basis set 3-21G determines the accuracy. From 3-21G to 6-31G(d,p)and 6-311+G(d,p). the accuracy increases. Here we are just using the lowest accuracy calculation set, bue the calculation time is quite short. The optimum position of the nuclei for a given electronic configuration is determined and it would give optimisation information of this molecule in a summary box as shown.
B-H=1.19 Å
H-B-H=120.0 deg
file type: .log
calculation type: FOPT
calculation method: RB3LYP
basis set: 3-21G
final energy: -69477kJ/mol, -26.46 a.u.
gradient: 0.00004507
dipole moment: 0.00D
point group:D3H
Job cpu time: 10.0sec
BCl3
find information about BCl3
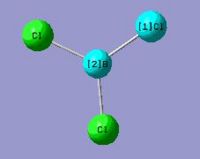
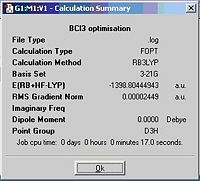
A BCl3 molecule is bulit up the same way as described for BH3. The calculation time much longer compare with BH3 even using the same caclculation method as Cl atom is much larger than H atom. Information is given in the summary box.
B-Cl= 1.78A
Cl-B-Cl Angle= 120.0 deg
File type: .log
Calculation type: FOPT
Calculation Method: RB3LYP
Basis Set: 3-21G
E(RB+HF-LYP): -3672561kJ/mol, -1398.80 a.u.
Dipole moment: 0.00 Debye
Point group: D3H
Job cpu time: 17.0 sec
vibrational analysis
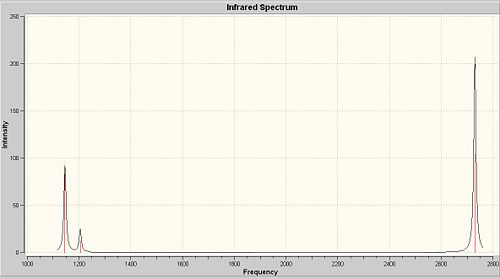
The Gaussian Output Files of optimised BH3 molecule is the starting point of its vibrational analysis. The job type of Gaussian Calculation is "frequency" this time instead of "optimisation". The Results of "Vibrations" will be shown in the "Results" menu.
The frequency analysis is the second derivative of the potential energy surface. If one of them is negative, a transition state is obtained. As seen all of six vibrations are positive, so the optimised molecule is sitting at the minized energy state as we expect. There are only three discrete peaks shown on the spectrum. Number 2 and 3 vibrations have exactly the same frequency, so they correspond to two degenerate orbitals and they will overlap to give a sharp peak. The reason is the same for vibrations 5 and 6. This is confirmed by the MO diagram. Vibration 4 has zero intensity, so it would not show any peak.
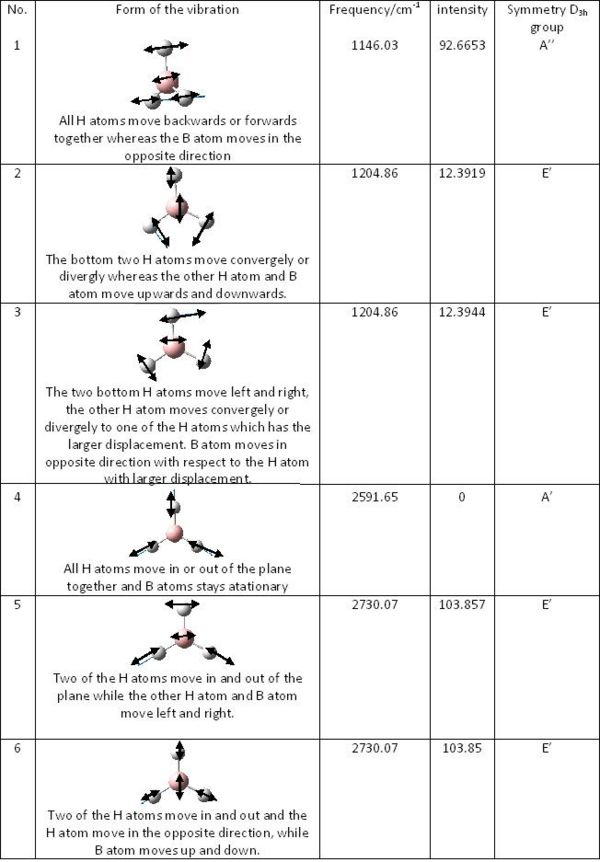
The results of the frequency analysis for BH3 are shown in the table:
molecular orbitals
Using Chemdraw, the predicted quanlitative MO diagram of trigonal planar D3h BH3 can drawn.

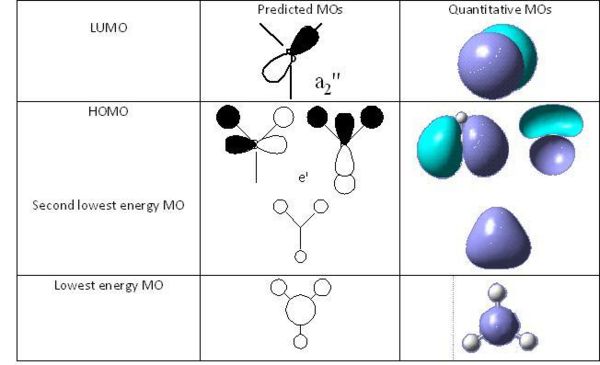
Now the predicted quanlitative and quantitative MOs at the same energy levels can be compared. By comparing the predicted qualitative and quantitative MOs, it can be seen that the qualitative MO shows very similar results as predicted qulitative MOs. SO qulitative MO theory is very accurate and useful.
H2O
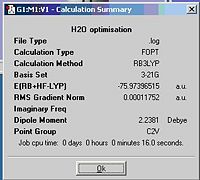
Small molecule H2O is chosen to take the optimisation, the method of calculation is the same as previous molecules. The following information is given in the resulting file. As H2O is larger than BH3, time taken to optimise it is longer when using the same calculation method.
the xyz coordinates of H2O:
Center Atomic Atomic Coordinates (Angstroms) Number Number Type X Y Z --------------------------------------------------------------------- 1 8 0 0.000000 0.000000 0.122755 2 1 0 0.000000 0.785462 -0.491020 3 1 0 0.000000 -0.785462 -0.491020
O-H bond distances: 1.00 A
H-O-H angle: 38.0 deg
calculation method: RB3LYP
basis set: 3-21G
final energy: -199470kJ/mol, -75.97 a.u.
dipole moment: 2.2381 D
point group; C2V
Job cpu time: 16.0 sec
Isomers of Mo(CO)4L2
To predict their thermal stabilities and spectral characteristics of the large molecules with better accuracy of opyimisation, like cis and trans isomers of Mo(CO)4L2 where L=P(CH3)3, a few steps are needed.
1. B3LYP/LanL2MB method is used for a rough optimisation first. Loose convergence criteria are also set. Convergence here relates to the first derivative of the energy.
2.The ground state optimised geometry is then optimised again using the method of B3LYP/LanL2DZ with increased convergence to obtain a better optimisation.
3. The IR frequencies of the cis and trans isomers then can be computed through SCAN system.
The predicted values and experiemtal values are compared in the tables below. It can be seen that the optimised and experimental values are very similar. The difference of bond distances is within 1A and of bond angle is within 1deg. Hence, the 2 steps optimisation gives quite accurate results.
comparsion of predicted bond distances of cis-Mo(CO)4(P(CH3)3)2 and the experimental values
| Bond | predicted bond distance/A | Experimental bond distance/A |
|---|---|---|
| Mo-P(10) | 2.65 | 2.525 |
| Mo-P(11) | 2.65 | 2.533 |
| Mo-C(2) | 2.03 | 1.997 |
| Mo-C(3) | 1.98 | 1.981 |
| Mo-C(4) | 2.03 | 2.034 |
| Mo-C(5) | 1.98 | 1.983 |
| C(2)-O(7) | 1.19 | 1.150 |
| C(3)-O(6) | 1.19 | 1.163 |
| C(4)-O(9) | 1.19 | 1.150 |
| C(5)-O(8) | 1.19 | 1.125 |
| P(10)-C(12) | 1.89 | 1.819 |
| P(10)-C(16) | 1.89 | 1.837 |
| P(10)-C(20) | 1.90 | 1.862 |
| P(11)-C(24) | 1.89 | 1.836 |
| P(11)-C(28) | 1.89 | 1.863 |
| P(11)-C(32) | 1.89 | 1.815 |
comparsion of predicted bond angles of cis-Mo(CO)4(P(CH3)3)2 and the experimental values
| Angle | predicted bond angle/deg | Experimental bond angle/deg |
|---|---|---|
| P(10)-Mo-P(11) | 95.3 | 94.75 |
| P(10)-Mo-C(2) | 89.8 | 87.8 |
| P(10)-Mo-C(3) | 86.2 | 89.3 |
| P(10)-Mo-C(4) | 89.6 | 87.1 |
| P(10)-Mo-C(5) | 175.2 | 175.5 |
| P(11)-Mo-C(2) | 88.7 | 87.8 |
| P(11)-Mo-C(3) | 178.5 | 176.3 |
| P(11)-Mo-C(4) | 88.9 | 89.7 |
| P(11)-Mo-C(5) | 89.5 | 94.0 |
| C(2)-Mo-C(3) | 91.2 | 89.9 |
| C(2)-Mo-C(4) | 177.4 | 177.0 |
| C(2)-Mo-C(5) | 90.4 | 89.2 |
| C(3)-Mo-C(4) | 91.2 | 92.9 |
| C(3)-Mo-C(5) | 89.0 | 88.4 |
| C(4)-Mo-C(5) | 90.4 | 89.9 |
| Mo-C(2)-O(7) | 179.4 | 178.0 |
| Mo-C(3)-O(6) | 179.8 | 179.1 |
| Mo-C(4)-O(9) | 179.4 | 178.5 |
| Mo-C(5)-O(8) | 178.9 | 178.0 |
The trans-isomer can be optimised in the same way.
comparsion of the optimised bond distances of trans-Mo(CO)4(P(CH3)3)2 and the experimental values
| Bond | Optimised bond distance/A | Experimental bond distance/A |
|---|---|---|
| Mo-P(10) | 2.57 | 2.4584 |
| Mo-C(2) | 2.03 | 2.024 |
| Mo-C(3) | 2.02857 | 2.035 |
| C(2)-O(6) | 1.19 | 1.144 |
| C(3)-O(7) | 1.19 | 1.135 |
| P(10)-C(12) | 1.89 | 1.840 |
| P(10)-C(16) | 1.89 | 1.826 |
comparsion of the optimised bond angles of trans-Mo(CO)4(P(CH3)3)2 and the experimental values
| Bond | Optimised bond angle/deg | Experimental bond angle/deg |
|---|---|---|
| P(10)-Mo-C(2) | 89.0 | 89.26 |
| P(10)-Mo-C(4) | 88.9 | 88.34 |
| Mo-C(2)-O(6) | 179.7 | 179.2 |
| Mo-C(3)-O(7) | 179.7 | 179.0 |
| Mo-P(10)-C(12) | 115.7 | 119.77 |
| Mo-P(10)-C(16) | 117.1 | 113.95 |
Stereochemistry of transition metal Mo carbonyl complexes can assigned by the CO stretching frequencies in the IR spectra. Disubstituted Mo tetracarbonyl octahedral complexes exhibit both cis and trans isomers. For non-linear molecules, there are 3n-6 vibrations, which is the case we are considering. The CO stretches usually occur in the range 2100-1750cm-1. However, the M-C stretches and M-C-O bends are normally found around 300 and 700cm-1. Overall, there will only be four CO stretching considered in the IR region. Before deciding the stereochemistry of products from IR spectra, point groups of cis and trans isomers of metal complexes will be discussed first.
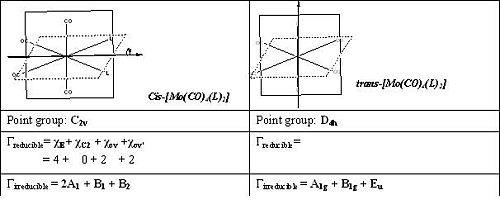
Compare the irreducible representation of cis and trans isomers, we find that all four components in cis-isomer are IR active whereas Eu is the only one active in trans isomer. [1]Thus, four CO absorption would be expected for cis-[Mo(CO)4(L)2] and only one would be expected for trans isomer.
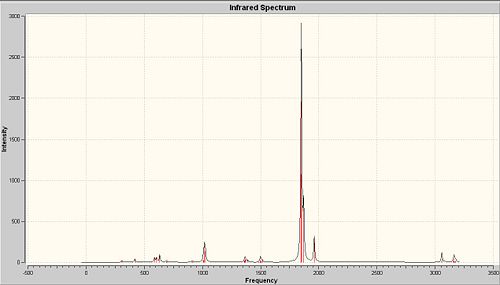
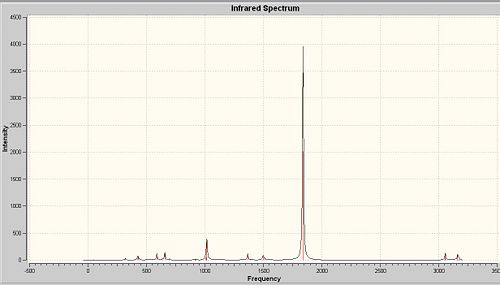
As seen from the calculated IR spectra of both isomers, the prediction about CO stretches is confirmed.
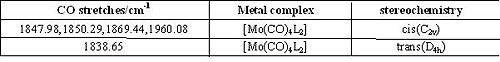
Compare with the experimental values of CO stretches obtained in second year lab, the predicted IR spectra show quite high accuracy, the CO stretches are shited about 50cm-1

Comparing the Cis and Trans Isomers
The Mo-P bond lengths of the trans-isomer (2.57A) is shorter than those of the cis-isomers (2.65A). In the cis-isomer, the Mo-C bond lengths of the carbonyl ligands(2.03A) are longer than those trans to the P(CH3)3 ligands (1.98A). This is due to the because the π-acceptors carbonyl ligands than the P(CH3)3 ligands.
comparsion of energies
| Cis isomer | Trans isomer | |
|---|---|---|
| Energy/a.u. | -773.36024012 | -773.35731208 |
| Energy/kJ/mol | -2030457 | -2030449 |
The cis-isomer is 8kJ/mol lower in energy than the trans-isomer, showing that the cis-isomer is thermodynamically slightly more stable. This may due electronic factors. However, cis-isomer is destablised by steric hindernce from L groups. Overall, the electronic factors affect the stability more sufficiently.stabilisation.
Ways to Alter the Relative Ordering of the Cis and Trans Isomers
Most of disubstituted Mo tetracarbonyl octahedral complexes would have a more stable trans isomer. The equilibrium would towards right hand side due to the greater rate constant of cis-trans formation than trans-cis formation. For the isomerisation of [Mo (CO)4L2], where L is a very bulky group, the cis isomer rearranges to the trans isomer via a dissociative mechanism. Mo-P would break first and a five-coordinate intermediate of [Mo(CO)4L2] would form. This intermediate would have the shape of square pyramidal with a L group in equatorial position. Another way is taking the attractive cation-π or NH-π interaction which can be used to force the equilibrium to the side with the cis-isomer. Also intermolecular hydrogen bonding can be introduced into the ligands L so that one of the atom in L that can hydrogen bond to the other ligand, forcing the ligands to be cis to each other.
Reference
1. M.Y.Darensbourg and D.J.Darensbourg, J.Chem.Ed., 1970(47),33
2. D.J.Darensbourg and R.L.Kemp,Inorg.Chem., 1978(17), 2680
3. A.D.Allen and P.F.Barrett,Can.J.Chem., 1968(46), 1649
4. D.J.Daresbourg,Inorg.Chem.,' 1979(18), 14
5. Yawen Zhang, Second Year Synthesis Lab Report S5, Identification of Stereochemical isomers of [Mo (CO)4(L)2] by infrared Spectroscopy
6. The molecules are optimised for a rough geometry using the B3LYP method and LanL2MB basis set.
The optimisations are deposited in the chemical database SCAN.
Unique identifier : 13175, 13177
7. Using the LanL2DZ/B3LYP method to optimise the geometry again at higher level.
Unique identifier : 13286-13287
8. The IR frequencies are then computed.
Unique identifier : 13377-13378
NH3
A low potential barrier exists between two inverted structures of ammonia by the hydrogen atoms quantum tunnelling from one side of the nitrogen atom to the other side.
symmetry and its impact
Different strctures of NH3 will be optimised using B3LYP/6-31G method to investigate the effect of symmetry on the structures, energies.
The calculation summary of the ground state of NH3 is shown below.
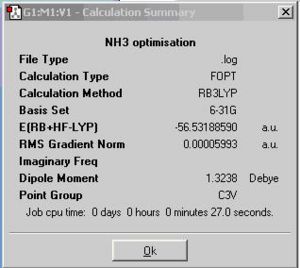
By altering on bond length into 1.01A, the symmetry of NH3 would change from C3v into C1
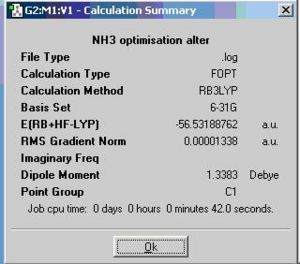
The calculation summary of the isomer of the planar inversion transition state NH3 is shown below.
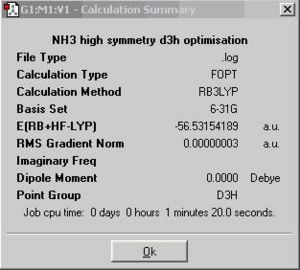
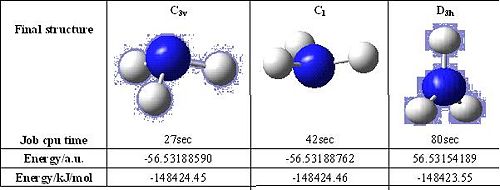
The C3v NH3 has a trigonal pyramidal structure with equal bond length and H-N-H bond angle(116.3°). The C1 NH3 has a distorted trigonal pyramidal structure with the three slightly different N-H bond lengths and H-N-H bond angles. The symmetry is broken. The D3h NH3 a trigonal planar structure with all the N-H bond lengths and H-N-H bond angles (120.000°) same.
The time taken to optimise the geometry of D3h NH3 is the much longest than the other two, following by the C1 and C3v NH3. C1 is a sub-group of C3v and C3v is a sub-group of D3h. So NH3 with point group D3h would have the most symmetry oprations. By looking at the structures of NH3, it can be seen that a molecule cannot "break symmetry" during an optimisation. If a high symmetry structure is entered, the symmetry with the best optimisation may not be able to be produced for the final geometry but this is useful when a geometry with a particular symmetry is aimed to be produced.
The D3h NH3 has the lowest energy geometry,it is higher than other two structures for 0.9 kJ/mol, which is not significant at all. The three NH3 molecules do not have significant difference, only the relative positions of H atoms are shifted. As H atom is smallest atom, so it would not alter the energy a lot.
the effect of using different methods
The effect of using a better method and baisis set is going to be investigated.
A NH3 molecule is created in the same way as before and optimised using MP2/6-311+G(d,p)method, instead of B3LYP/6-31G. This produces the ground state C3v NH3 and the planar inversion transition state which has D3h symmetry.
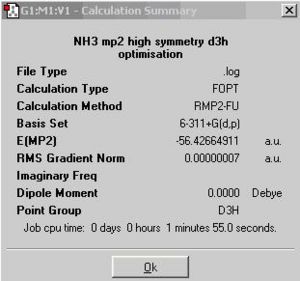
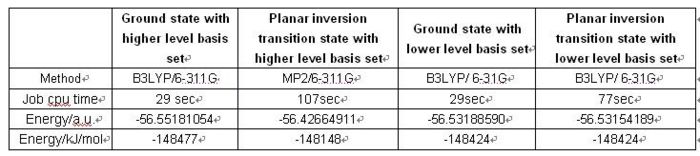
There is no effect on job cpu time when changing the basis set from 6-31G to 6-311G for ground state calculation. But the method and basis set changing changes the energy difference between ground state and transition state a lot. The lower basis set does not show any difference in energy between ground and transition state. The barrier height to inversion at higher basis set is 329kJ/mol, which is almost 14 times larger than the experimental value 24.3 kJ/mol. A higher level basis set may be used to obtain a better result.
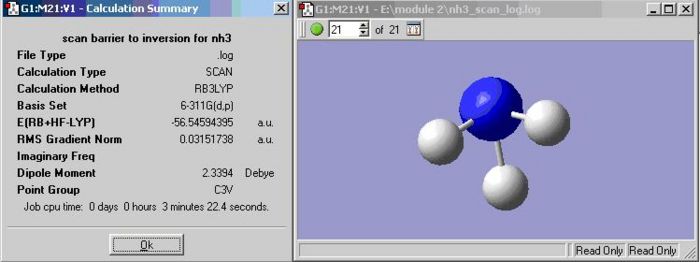
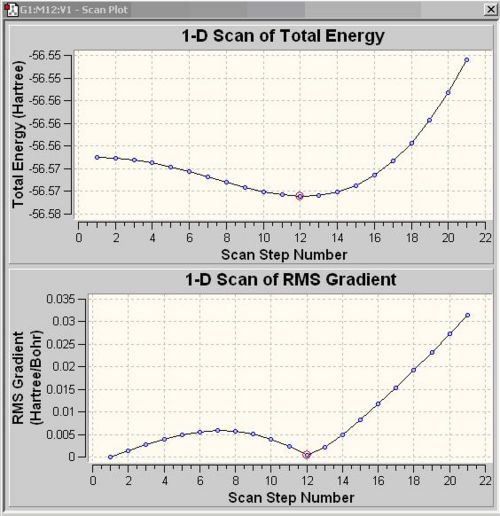
following the inversion path
In order to view the structures between ground state and transition state together on a "reaction path", "scan" has to be computed by varying one coordinate while allowing all others to optimise. For the NH3, the H atoms are set to move down from the plane of the N atom. The number in the box next to the green buttom indicates the number of energy states, there are 21 states for NH3. Plots show the energy and the gradient of energy at each step of the optimisation and structures of the molecule at each step. The PES scan shows NH3 achieves the lowest energy at scan step 12. The Root Mean Square Gradient curve shows the gradient going to zero as the minimum is approached.
the vibrational connection
There is no negative frequency of C3v NH3 whereas there is one in D3H NH3. There are six frequencies for each NH3 molecule. The only negative frequency at -318.05cm-1 of D3H NH3 indicates that it is transition state structure. Vibration number 2 in the C3v and D3h structures follows the inversion reaction path.

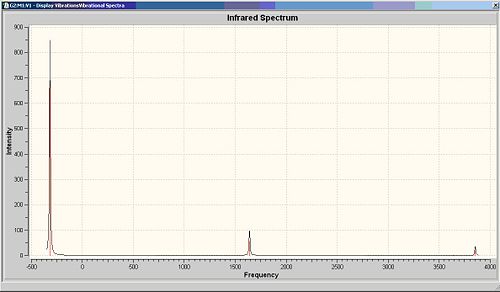

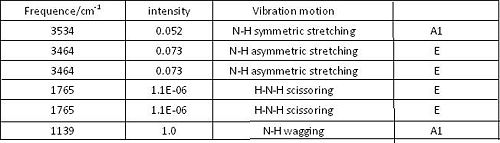
Compare the calculated IR frequencies with literture values, these two sets of data show similarity for vibrations 2-6. The vibration with lowest frequency is quite different. This may be improved by using a higher accuracy calculation method.
Reference
1. http://www.chem.purdue.edu/gchelp/vibs/nh3.html
Mini Project
MoOCl2(PR3)3 could be isolated in both blue and green isomeric forms. The original thought about this isolation is due to the difference between cis and trans isomers of this complex. But further research indicates that both green and blue forms are from cis-isomer, and the difference is actually due to the distortional isomers of cis-isomer. They are called mer-isomers. In this mini project, cis and trans-MoOCl2(PMe3)3,cis-WOCl2(PMe3)3 and cis-MoOCl2(PMe3)2(NMe3will be investigated into. The key point is to look at the Mo-O bond, including bond length and IR stretch to investigate which form of this cis-MoOCl2(PMe3)3 would modelling by Gassview. Firstly the general electronic structures of these complexes will be looked at.
The inital molecule that is going to be investigated is cis-MoOCl2(PMe3)3. This was drawn in guassview. The molecule was initally drawn in octahedral shape, then a DFT/B3LYP/3-21G optimisation(as the complexes are quite large,only basic calculation methos is used) and frequency calculation was run through SCAN system.
The resulting cis-MoOCl2(PR3)3 molecule can be seen below:
Pentahelicene |
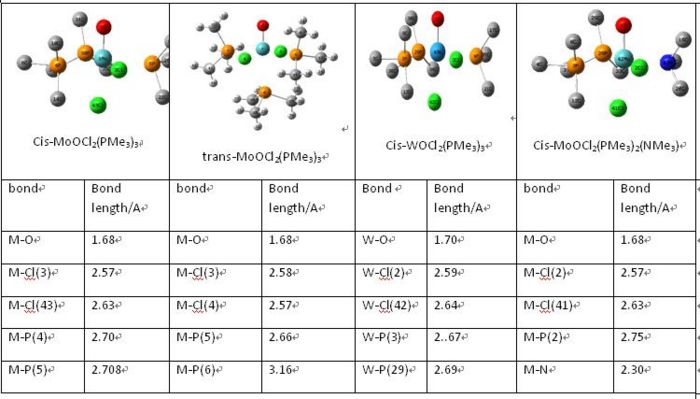

The value of M-O bond is 1.68A for Mo-O and 1.70A for W-O bond. The metal-O bond would then be considered as M=O bond. These values are very close to the one set of experimental results. There are more than one from of each molecule with silghtly different bond lengths of M-O and M-Cl. The comparison of bond length indicates that the computational model only exits in one form. Other bond lengths of cis-MoOCl2(PMe3)3 are slight greater in calculated results, generally 0.2A greater. By looking at the Mo-N, it can been seen that this Mo-N bond is much shorter than Mo-P. So the Mo-N is stronger due to the stronger π donor NMe. This would alter the MO distribution as well.
The next thing that is to look at is the generated vibration of the molecule. M-O stretch is likely to be found in the region of 930-980cm-1. This stretch is observed in all complexes as a small peak at around 950-1 next to the sharp C-H stretch.
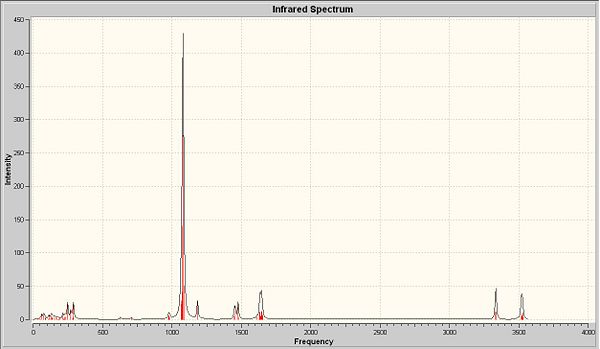
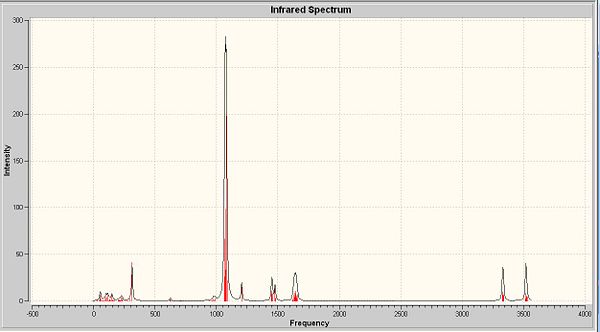

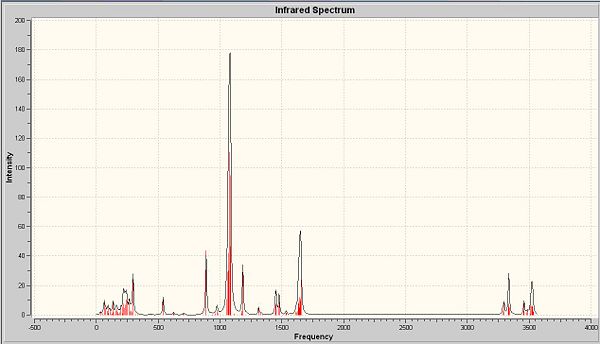
There are two crystallographically independent molecules of cis-MoOCl2(PMe3)3 and the Mo-O bond lengths are different in these two isomers. They are crystallographically inequivalent, but chemically equvilant. Summary above, there should be two stretching frequencies of Mo-O. But in the predicted and experimental IR, only one stretching frequency is observed. This is also applied to the experimental results due to the presence of mixtures of two-mer-isomers. So the computational model is in the between of two-mer-isomers. With the data of bond length, the major isomer is the one with shorter M-O length. Among these four complexes, the cis-MoOCl2(PMe3)2(NMe3) shows the biggest difference with respect to the other three.
The stretch at 1654cm-1 indicated the presence of C-N, which is absent from the other three complexes. There are two M-Cl stretches at around 350cm-1 in each complexes.
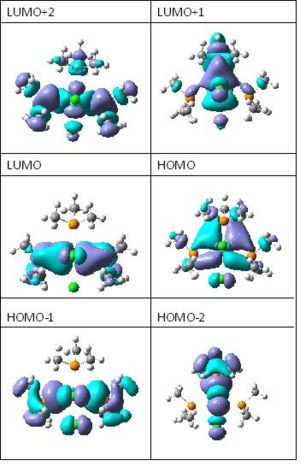
The next thing that is look at is the generated MO's of the molecules cis-MoOCl2(PMe3)3 and cis-MoOCl2(PMe3)2(NMe3. NMe is a strong π-donor, the interaction between N and d orbital of Metal is much stronger. So the electrons are more densed. This can be clearly seen from the snapshot of LUMO an HOMO of both complexes.
Here are the molecular orbitals of both complexes:

FInally, it is to look at the energy of these four complexes.cis and trans-MoOCl2(PMe3)3 have similar energy and they are the highest compare the rest. cis-MoOCl2(PMe3)2(NMe3) has much lower energy than the others, as it is stablilsed by the stronger interaction between metal and N donor.
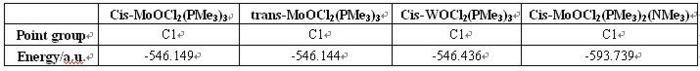
This part of the project cannot clearly display all forms of cis-MoOCl2(PMe3)3 with two different colours. Futher study on NMR and UV-Vis may explain more. But as seen from the comparison of the four similar complexes, the property of subsititude can alter the properties of the complex whereas the central metal in the same group would not change a lot.
Reference:
1. The optimisations are deposited in the chemical database SCAN.
Unique identifier for the these four complexes : 13371-13374
2. The frequencies are deposited in the chemical database SCAN.
Unique identifier for the these four complexes : 13416-13417, 13426-13427
3. The MO analysis is deposited in the chemical database SCAN.
Unique identifier : 13711-13712
4. M.J.Morris, Coordination Chemistry Reviews., 1996(152), 309-358
5. G. Parkin, Chem. Rev., 1993, 93(3), 887-911, DOI:10.1021/cr00019a003
