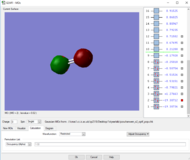
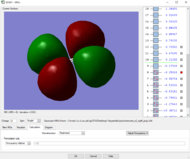
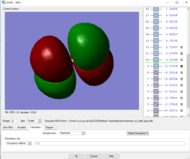
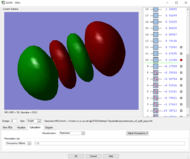
Rep:Mod:zp2518
NH3 molecule
summary
Calculation MethodːRB3LYP
Basis Setː6-31G(d,p)
Final Energy(RB3LYP)ː-56.5577687 a.u.
RMS Gradientː0.0000049 a.u.
Point GroupːC3V
N-H bond distance = 1.02 Å
H-N-H bond angle = 106°
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
The maximum force is below 0.00045 and the converge of the set of values means that the optimisation is completed
JMol of structure
NH3 |
The optimisation file is linked here
Display vibrations table
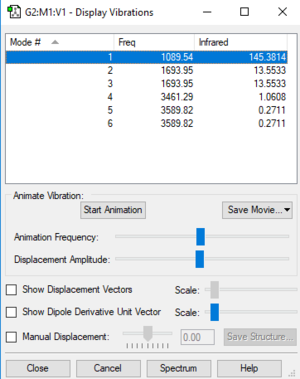
Table of vibrations and intensities
| wave number cm-1 | 1090 | 1694 | 3461 | 3590 |
| symmetry | A1 | E | A1 | E |
| intensity arbitrary unit | 145 | 14 | 1 | 0 |
| image |  |
 |
 |
|
The vibrations at 1694 cm-1 and 3590 cm-1 have two degenerate states respectively, 6 modes of vibrations in total
The difference in the intensity is due to the different in the change of dipole during the vibration . The first mode have the highest intensity among the 6 modes because it has the highest change in the dipole (all H-atoms move to the same side). And the two degenerate vibration, modes 5&6, have the lowest change in dipole among the 6 modes hence the lowest intensity.
Question and answer
how many modes do you expect from the 3N-6 rule?
6.
which modes are degenerate (ie have the same energy)?
Modes2&3 , modes5&6.
which modes are "bending" vibrations and which are "bond stretch" vibrations?
Modes1,2,3 are bending. Modes4,5,6 are bond stretch.
which mode is highly symmetric?
Modes 1,4 are highly symmetric.
one mode is known as the "umbrella" mode, which one is this?
Mode 1.
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
We expect to see four bands, because modes 2&3 and 5&6 are degenerate. But two of the four bands are too low in intensity (1.06&0.27) to be visualise in an real experiment due to the limitation of experimental appliance.
NBO charge
Charge on the N-atom = -1.125
charge on the H-atom = 0.375
The N-atom are expected to have the negative charge due to its high electronegativity
H2 molecule
summary
Calculation MethodːRB3LYP
Basis Setː6-31G(d,p)
Final Energy(RB3LYP)ː-1.1785394 a.u.
RMS Gradientː0.0000002 a.u.
Point GroupːD∞h
H-H bond distance = 0.74 Å
linear structure
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
The maximum force is below 0.00045 and the converge of the set of values means that the optimisation is completed
JMol of structure
H2 |
The optimisation file is linked here
Display vibrations table
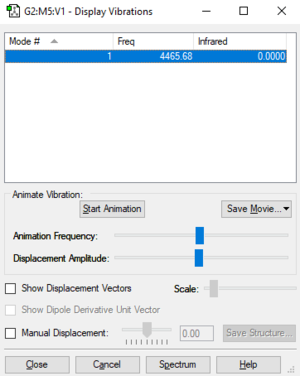
Table of vibrations and intensities
| wave number cm-1 | 4466 |
| symmetry | SGG |
| intensity arbitrary unit | 0 |
| image | 
|
The intensity of this vibration is zero due to the vibration is symmetric and there's no change in the dipole.
NBO charge
Charge on both of the H-atoms = 0
The two H-atoms have same electronegativity, the charges are cancelled out due to the molecule's symmetry, so the NBO charges are 0.
N2 molecule
summary
Calculation MethodːRB3LYP
Basis Setː6-31G(d,p)
Final Energy(RB3LYP)ː-109.5241287 a.u.
RMS Gradientː0.0000006 a.u.
Point GroupːD∞h
N-N bond distance = 1.11 Å
Linear structure
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
The maximum force is below 0.00045 and the converge of the set of values means that the optimisation is completed
JMol of structure
N2 |
The optimisation file is linked here
Display vibrations table
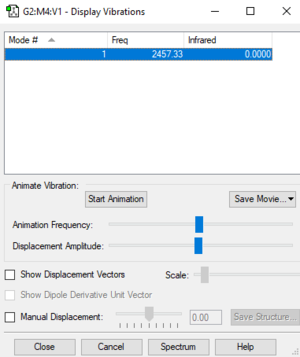
Table of vibrations and intensities
| wave number cm-1 | 2457 |
| symmetry | A1 |
| intensity arbitrary unit | 0 |
| image | 
|
The intensity of this vibration is zero due to the vibration is symmetric and there's no change in the dipole.
NBO charge
Charge on both of the N-atoms = 0
The two N-atoms have same electronegativity, the charges are cancelled out due to the molecule's symmetry, so the NBO charges are 0.
Mono-metallic TM complex that coordinates H2
DECXOY
Sturcture displayed[here]
H-H Bond distance in the structure with identifier DECXOY = 0.9 Å
H-H bond distance gained by computational calculation = 0.74 Å
The computational distances H-H is smaller than that in the structure we gained(DECXOY) which means the H-H bond is shorter and stronger. This can be explained by the difference in the environment of the H-atoms. In the structure, H-atoms are bonded to metal ion and the electronegative TM pull the electron density away form the hydrogens. The electron density diffusion cause the strength of bond between the H-atoms in the structure to be lower, hence longer distance. In addition, in the computational calculation, only parameters are used, while in real experiment, environment should also be taken in to consideration. The effect of this computational failure could be also be an explanation to difference in the H-atoms distance.
Determining the energy for Haber-Bosch process
E(NH3)= -56.5577687 a.u.
2*E(NH3)= 3*-1.17853936 = -113.1155375 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= 3*-1.17853936 = -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
=-113.1155375-[-109.52412868+-3.53561808]
=-0.05579064 a.u.
=-146.5 KJ/mol
The energy change have negative sign, so the ammonia product is more stable.
O2 molecule
summary
Calculation MethodːRB3LYP
Basis Setː6-31G(d,p)
Final Energy(RB3LYP)ː-150.2574243 a.u.
RMS Gradientː0.0000750 a.u.
Point GroupːD∞h
O=O bond distance = 1.22 Å
linear structure
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000130 0.000450 YES
RMS Force 0.000130 0.000300 YES
Maximum Displacement 0.000080 0.001800 YES
RMS Displacement 0.000113 0.001200 YES
The maximum force is below 0.00045 and the converge of the set of values means that the optimisation is completed
JMol of structure
O2 |
The optimisation file is liked to here
Display vibrations table
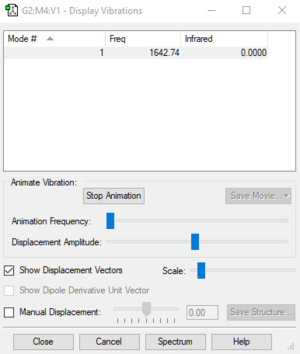
Table of vibrations and intensities
| wave number cm-1 | 1643 |
| symmetry | SGG |
| intensity arbitrary unit | 0 |
| image | 
|
The intensity of this vibration is zero due to the vibration is symmetric and there's no change in the dipole.
NBO charge
Charge on both of the O-atoms = 0
The two O-atoms have same electronegativity, the charges are cancelled out due to the molecule's symmetry, so the NBO charges are 0.
MOs
IndependenceːCO2 molecule
summary
Calculation MethodːRB3LYP
Basis Setː6-31G(d,p)
Final Energy(RB3LYP)ː-188.5809395 a.u.
RMS Gradientː 0.0000115 a.u.
Point GroupːD∞h
C=O bond distance = 1.17 Å
O=C=O bond angle = 180 °
linear structure
"Item" table
Item Value Threshold Converged?
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000015 0.001200 YES
The maximum force is below 0.00045 and the converge of the set of values means that the optimisation is completed
JMol of structure
CO2 |
The optimisation file is liked to here
Display vibrations table
Table of vibrations and intensities
| wave number cm-1 | 640 | 1372 | 2436 |
| symmetry | PIU | SGG | SGU |
| intensity arbitrary unit | 31 | 0 | 545 |
| image |  |
 |
|
The vibration at 640 cm-1 have two degenerate vibration states.
The intensity of second mode of vibration is zero due to the vibration is symmetric and there's no change in the dipole. The 4th mode of vibration has higher intensity than the 1st and 2nd ones' because the 4th mode vibration have higher change in the dipole.
NBO charge
Charges on both of the O-atoms and C-atom = 0
The two O-atoms have same electronegativity, the charges are cancelled out due to the molecule's symmetry as the three-atom molecule have linear shape, so the NBO charges are 0.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, very good explanations overall, well done!
One area I would improve is when a molecule is unoccupied you call it a non-bonding MO. It is clearer to say for e.g. the LUMO: This MO has antibonding interations between the O atoms, but does not reduce the bonding in the molecule because it is unnoccupied.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
You did an extra calculation, well done.
Do some deeper analysis on your results so far