Rep:Mod:yyl25012502
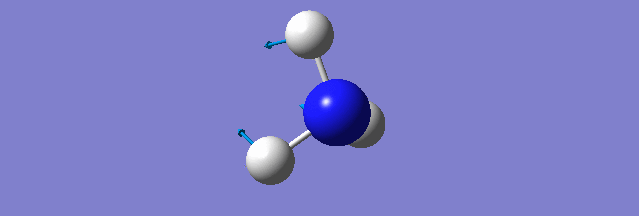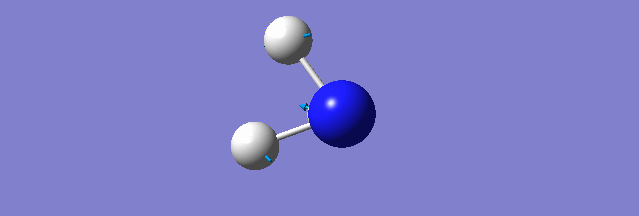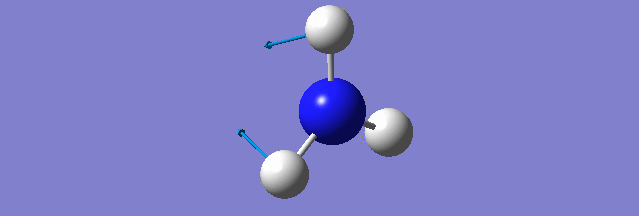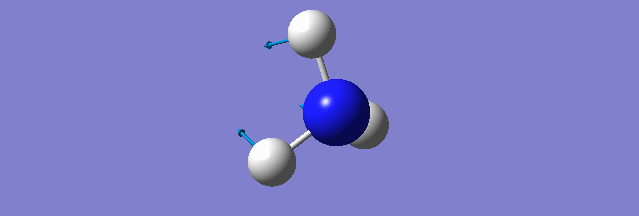
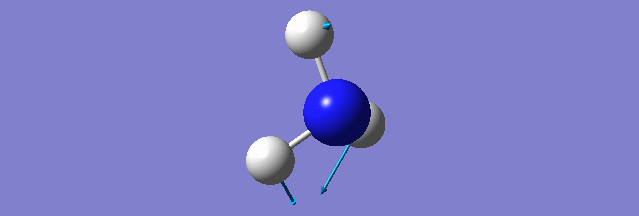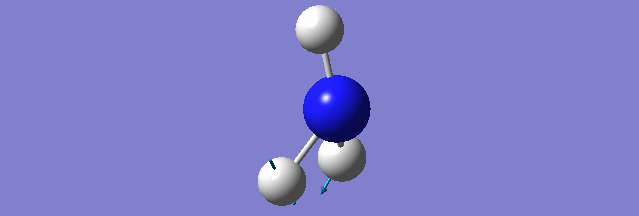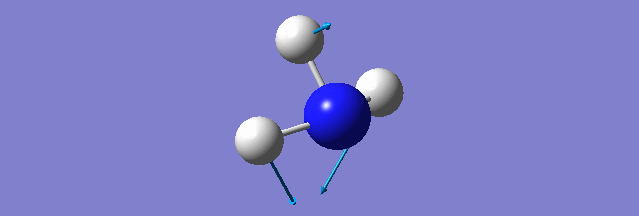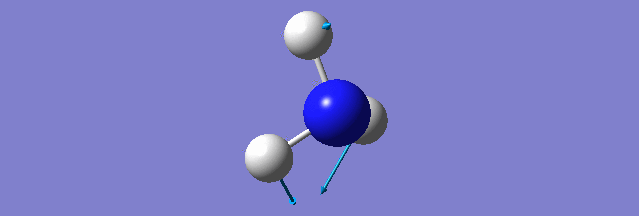
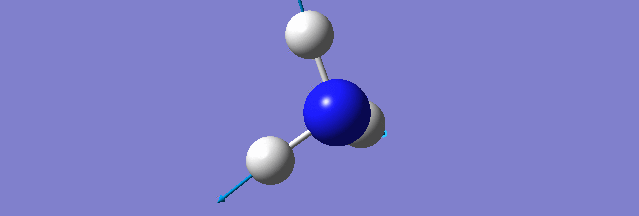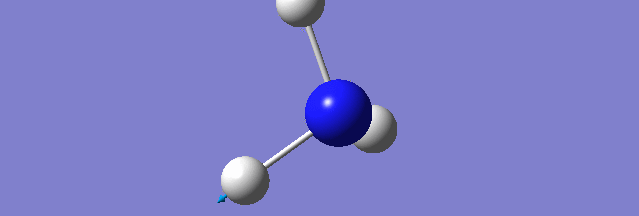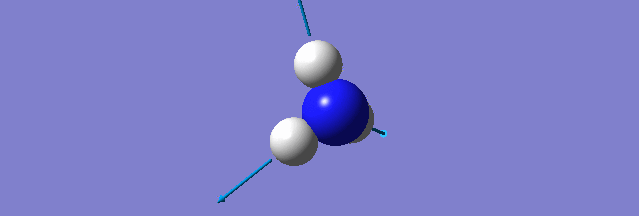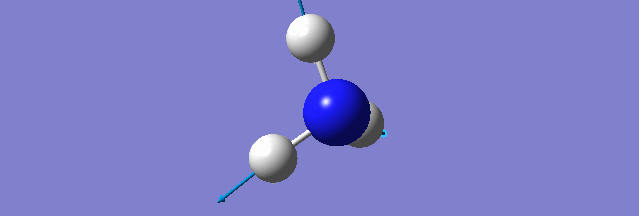
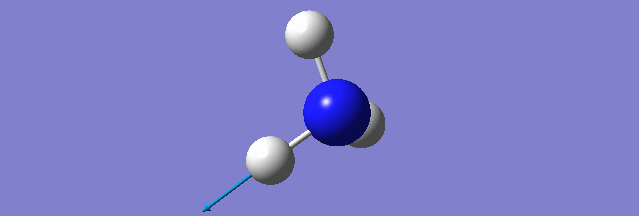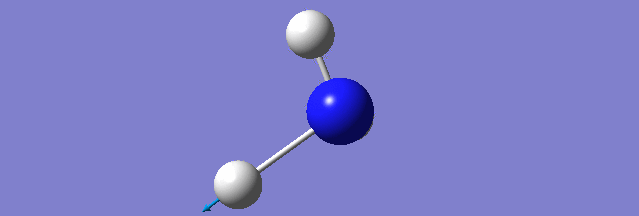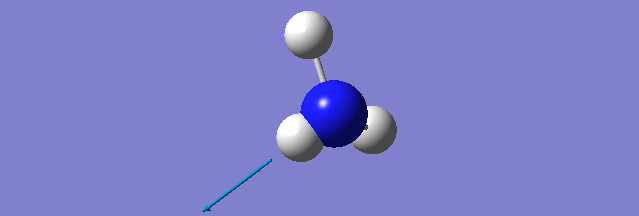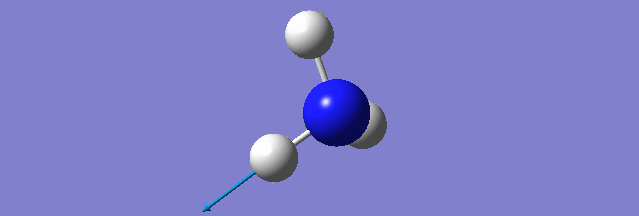
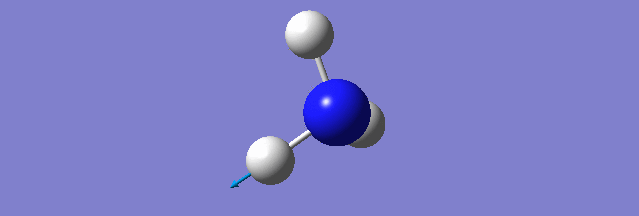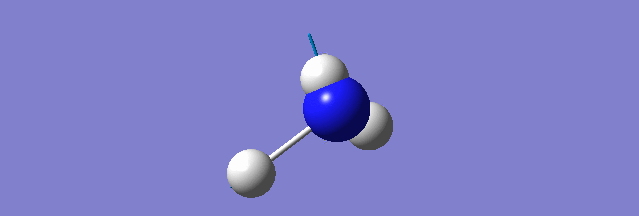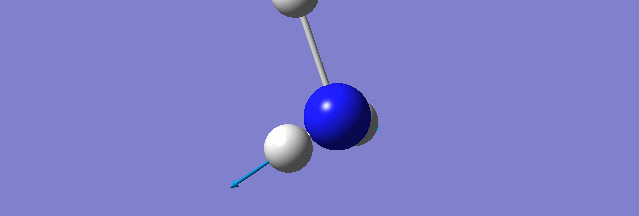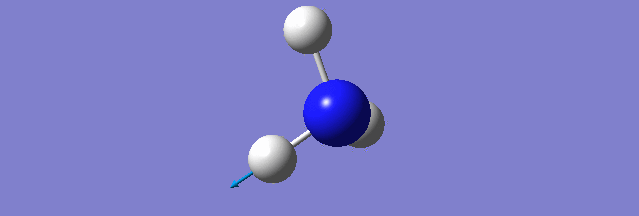
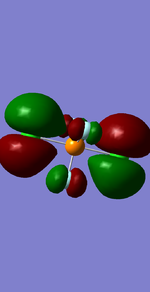
NH3
NH3 |
The optimisation file is linked to File:TINOLAM NH3 OPTIMISATION POP 10.LOG
Structure and Reactivity of NH3
Calculation method= RB3LYP The basis set= 6-31G(d.p) final energy in atomic units= -109.52359111 a.u. The RMS gradient= 0.02473091 The point group of the molecule= C3V N-H bond distance= 1.3 angstrom ≈ 0.01Å optimised H-N-H bond angle = 109.471 degree ≈ 1°
Vibration of molecule
There should be 6 modes of vibration derived from the 3N-6 rule. The 2nd and 3rd modes are degenerate as a set while the 5th and 6th are another degenerate set. The first,second and third modes are bending while the rest are bond stretching vibration, the 4th mode is highly symmetric. The first mode is found to be the umbrella mode. In an experimental spectrum approximately only 2 bands, which is at the range of 150 and 10 respectively, can be seen.
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Charges of Molecule
The charge of the N atom is found to be -1.125 while the NBO charge of the 3 hydrogen atoms are found to be 0.375.
N2
N2 |
The optimisation file is liked to File:TINOLAM N2 PLEASEDONTFAIL.LOG
Structure and Reactivity of N2
Calculation method= RB3LYP The basis set= 6-31G(d.p) final energy in atomic units= -56.44397188 a.u. The RMS gradient= 0.05399560 The point group of the molecule= D*H N-N bond length=1.09200 angstrom
Charges of the N2
NBO Charge of nitrogen atom is zero.
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Vibration of the molecule
| N-N mode of vibration | ||
|---|---|---|
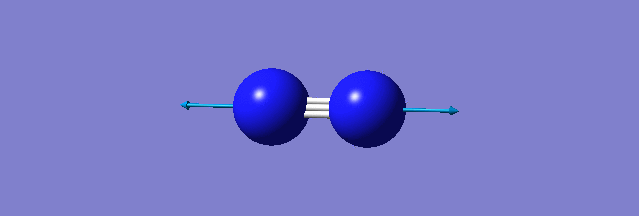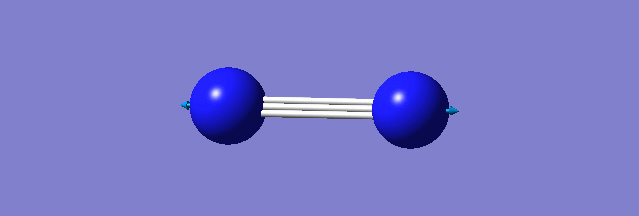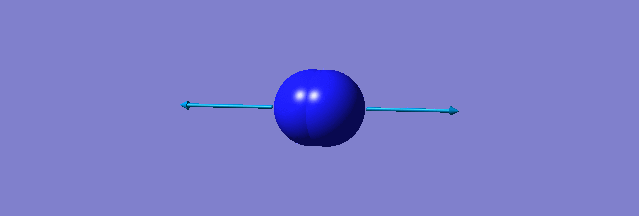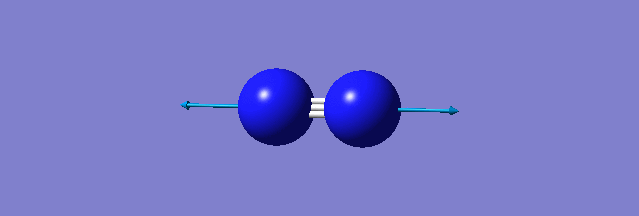
| ||
| wavenumber(cm-1) | 2457 | |
| symmetry | SGG | |
| intensity | 0.0000 | |
Comparism of structure of the molecule with other compound
From Conqeust App, I extrated the data from VEJDIA(P-1) which is one of the mono-metallic TS complex and the bond length of N-N is 1.105. The bond length is found to be longer than the single N2 molecule and the explanation of that is because of the delocaslised benzene ring in the compound that drag the electron density of N2 away and makes the bond longer.
H2
N2 |
The optimisation file is liked to File:TINOLAM H2 GOODTHING.LOG
Structure and Reactivity of H2
Calculation method= RB3LYP The basis set= 6-31G(d.p) final energy in atomic units= -1.15928020 a.u. The RMS gradient= 0.09719500 The point group of the molecule= D*H H-H bond distance= 0.6 angstrom ≈ 0.01Å
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Charges of the H2
NBO Charge of hydrogen atom is zero.
Vibration of the molecule
| H-H mode of vibration | ||
|---|---|---|

| ||
| wavenumber(cm-1) | 4465.68 | |
| symmetry | SGG | |
| intensity | 0.0000 | |
Haber-Bosch reaction energy calculation
E(NH3)= -109.5 au 2*E(NH3)= -219 au E(N2)= -56.4 au E(H2)= -1.2 au 3*E(H2)= -3.6 au ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -159 au
PCl2F3
NH3 |
The optimisation file is linked to File:TINOLAM PF2CL2 BANGER.LOG
Structure and Reactivity of PCl2F3
Calculation method= RB3LYP The basis set= 6-31G(d.p) final energy in atomic units= -1561.33841107 a.u. The RMS gradient= 0.01179626 The point group of the molecule= D3H N-H bond distance= 1.3 angstrom ≈ 0.01Å optimised H-N-H bond angle = 109.471 degree ≈ 1° P-Cl bond length= 2.04 angstrom. P-F bond length=1.63 angstrom. Cl-P-Cl bond angle= 180 degree. F-P-F bond angle = 120 degree.
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000046 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Vibration of the Molecule
| 1st Mode | ||
|---|---|---|
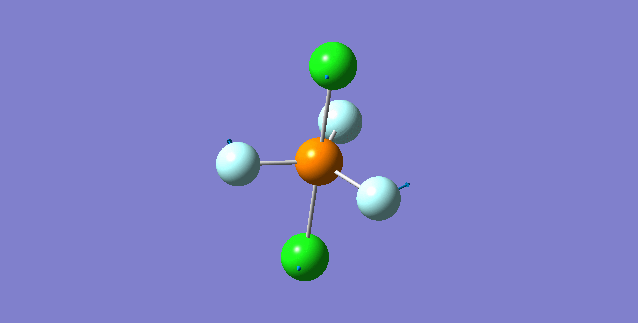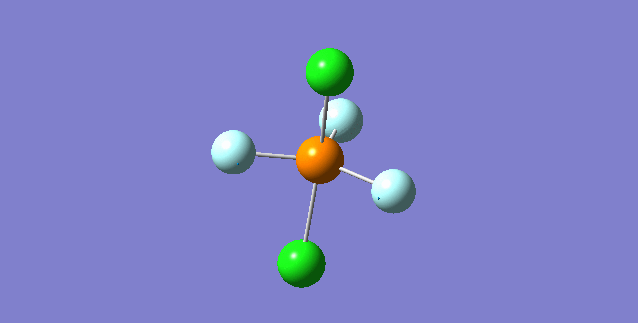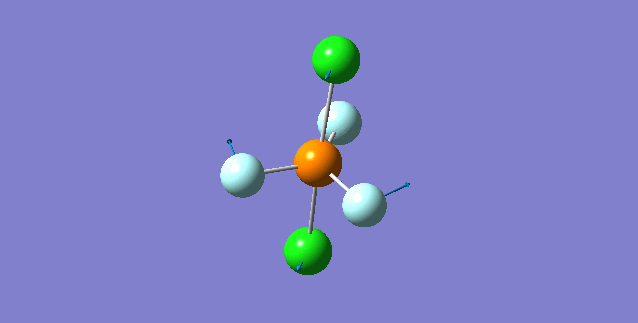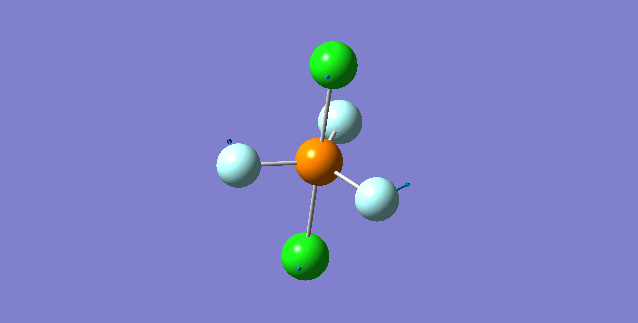
| ||
| wavenumber(cm-1) | 117 | |
| symmetry | E' | |
| intensity (a.u.) | 0 | |
| 2nd Mode | ||
| File:File:Super 2nd mode aofujhodhfsdfaasdf.gif | ||
| wavenumber(cm-1) | 117 | |
| symmetry | E' | |
| intensity (a.u.) | 0 | |
| 3rd Mode | ||
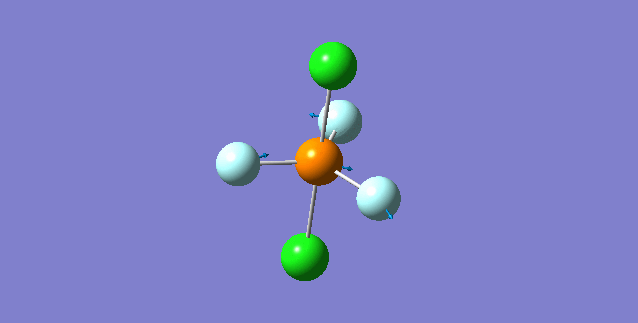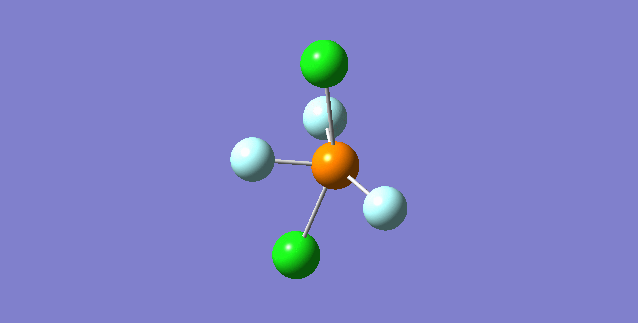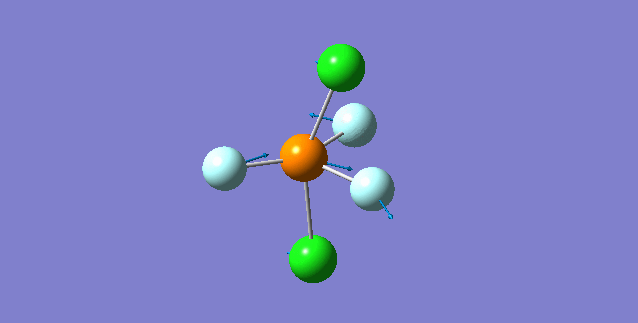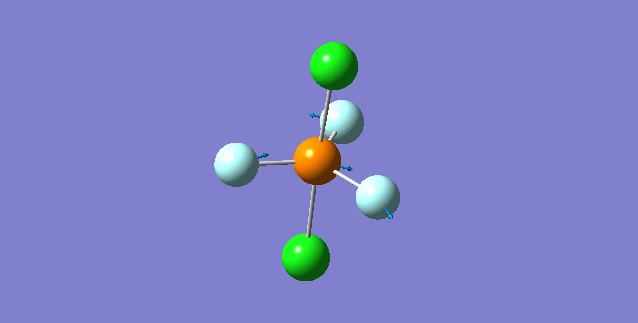
| ||
| wavenumber(cm-1) | 348 | |
| symmetry | A | |
| intensity (a.u.) | 13 | |
| 4th Mode | ||
| File:File:Super 4th mode awidfuaisudfia.gif | ||
| wavenumber(cm-1) | 348 | |
| symmetry | A | |
| intensity (a.u.) | 13 | |
| 5th Mode | ||
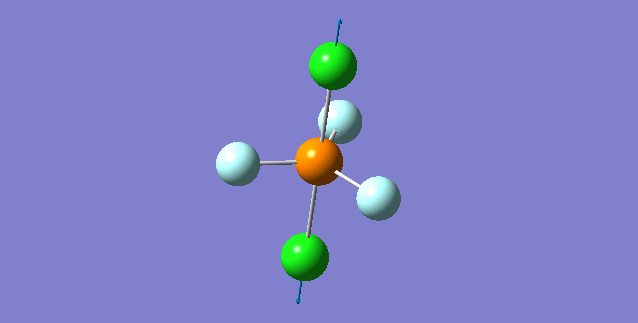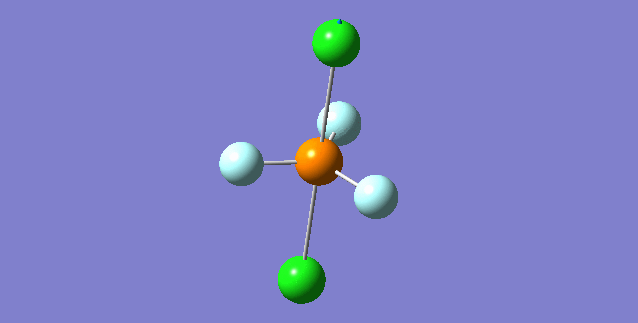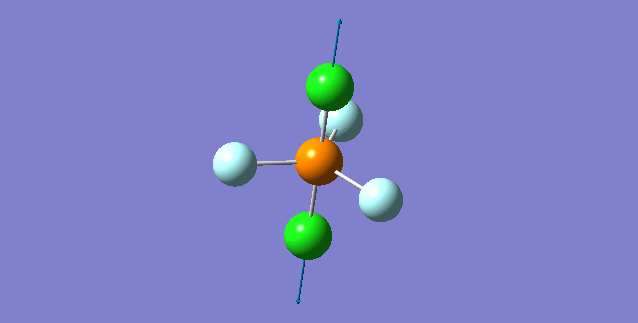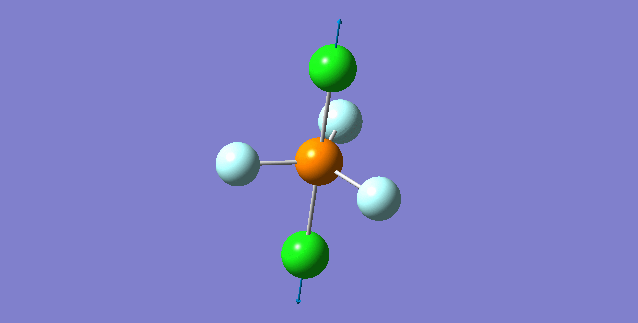
| ||
| wavenumber(cm-1) | 353 | |
| symmetry | A | |
| intensity (a.u.) | 0 | |
| 6th Mode | ||
| ||
| wavenumber(cm-1) | 371 | |
| symmetry | E" | |
| intensity (a.u.) | 0 | |
| 7th Mode | ||
| ||
| wavenumber(cm-1) | 371 | |
| symmetry | E" | |
| intensity (a.u.) | 0 | |
| 8th Mode | ||
| ||
| wavenumber(cm-1) | 451 | |
| symmetry | A2" | |
| intensity (a.u.) | 2 | |
| 9th Mode | ||
| ||
| wavenumber(cm-1) | 589 | |
| symmetry | A2" | |
| intensity (a.u.) | 683 | |
| 10th Mode | ||
| ||
| wavenumber(cm-1) | 707 | |
| symmetry | A1' | |
| intensity (a.u.) | 0 | |
| 11th Mode | ||
| ||
| wavenumber(cm-1) | 985 | |
| symmetry | E' | |
| intensity (a.u.) | 189 | |
| 12th Mode | ||
| ||
| wavenumber(cm-1) | 985 | |
| symmetry | E' | |
| intensity (a.u.) | 189 | |
NBO Charges
P=2.193 F=-0.528 Cl=-0.304
MO Diagram
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
You have accidentally measured the bond lengths from the unoptimised structure, and so have reported an incorrect value.
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES - you added animations well done!
Have you included answers to the questions about vibrations and charges in the lab script?
YES - good answers to the vibrations questions well done! You could have improved in your discussion of the charges by explaining the values obtained with reference to the electronegativities of the atoms.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES - you have made a good effort here. Unfortunately you have taken your bond lengths from the first step of the calculation, rather than the last step with the optimised structure. The jmol automatically shows the unoptimised first step of the .log file, you need to right click on it and select the final step to view the optimised structure.
Crystal structure comparison 0/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
You searched for and found a structure, well done. A link to the structure is missing however.
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 0/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
You have done the correct calculations well done! However you have used au units and you have rounded them too far. When doing calculations you should keep all sig figs until you come to the final answer and then round the answer appropriately, otherwise compounding errors result in an incorrect answer. e.g. if I calculate 1.04 x 7 I get 7.28, which I can round to 1 d.p. giving 7.3 (correct). If I round my values before doing the calculation I get 1 x 7 = 7.0. (incorrect)
Have you reported your answers to the correct number of decimal places?
You should have reported your final answer in Kj/mol, not au.
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
You missed out the question.
Your choice of small molecule 2/5
Have you completed the calculation and included all relevant information?
YES - all information is present well done. However you have left bits of copy/pasted NH3 data in this section also.
Have you added information about MOs and charges on atoms?
YES - you have added some images of MOs and values of the charges on atoms. To improve you could have explained the charges on the atoms using an electronegativity argument. You could have also explained the AO contributions to the MOs in more detail.
Independence 0/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far