Rep:Mod:yunzhang Module2 miniproject
Mini Project
Abstract
The reaction of copper chlorides with 5-methylfurfural thiosemicarbazone(M5FTSC) gives two series of new complexes: [M(M5FTSC)2X2]and [M(M5FTSC)X2]. In this case, the geomometric and the spectroscopic properties of [CuCl2(M5FTSC)]have been studies by using Gaussview 3.09.
| M5FTSC ligand | [Cu(M5FTSC)Cl2] complex | ||||||
|---|---|---|---|---|---|---|---|
|
The Cu atom is coordinated through the S atom and the azomethine N atom |
Optimisation
The structure of both M5FTSC ligand and [CuCl2(M5FTSC)] complex are optimised 1st under B3LYP/LANL2MB and further optimised under LANL2DZ.
| Name of molecule | Results Summary for the 1st optimisation | Results Summary for the 2nd optimisation |
|---|---|---|
| M5FTSC | 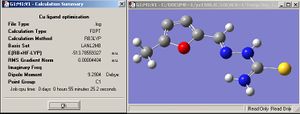 |
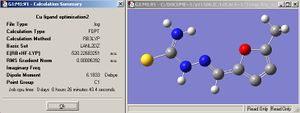 |
| [CuCl2(M5FTSC)] |  |
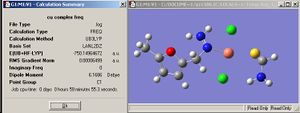 |
| M5FTSC ligand after 2nd optimisation | [Cu(M5FTSC)Cl2] complex after 2nd optimisation | ||||||
|---|---|---|---|---|---|---|---|
|
|
IR analysis
| IR spectrum for (M5FTSC)ligand | IR spectrum for [Cu(M5FTSC)Cl2] complex |
|---|---|
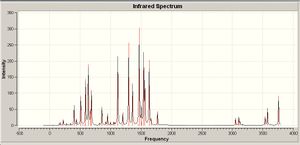 |
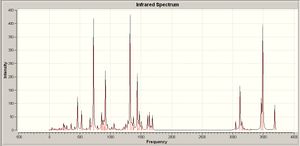 |
The calculated IR absorptions coorelate reasonably well with the literature value. The IR spectra for the ligand shows a strong band at aroung 1600cm-1 which corresponds to the C=N stretches. It is intersting to note that when ligand bonds to metal, all most all the other frequencies shift to a high region and the new band appears around 400 indicating the formation of the Cu-N bond.
DOI:
http://hdl.handle.net/10042/to-1920
http://hdl.handle.net/10042/to-1968
http://hdl.handle.net/10042/to-1940
http://hdl.handle.net/10042/to-1940
http://hdl.handle.net/10042/to-1973
Bond length and Bond angle compare to literature value for both ligand and complex
Bond length

Bond angle

The calculated bond distances and angles for the ligand coorelate well to the literature values, however this is not the case for the complex. The values highlighted in pink is the ones correlate poorly. It is most noticeable that the S-C1 and N2-C1 bond lengths are differ dramatically form the literature value, this might due to the programme draws bonds based on a distance critera such as how close the atoms are and how they interact. Here, the distance of bonds exceed some pre-defined value and some bonds in the optimised structure dissociate, therefore the difference between these two bond lengths are large.
Conclusion
Based on the optimisation the [CuCl2(M5FTSC)] complex and the M5FTSC ligand, their geomometric and the spectroscopic properties of [CuCl2(M5FTSC)]have been analysed and compared with literature value and ingeneral, there is a good coorelation between the calculated and the literature values for them. The simplier the molecule is, the better the optimisation can be carried out using the GaussView program and the less expensive in terms of time it is. However, for large and complex molecules, a higher level of optimisation is needed in order to obtain an optimum calcualted result. This programme is good in terms of visualising the atom coordination and the bond stretching of simple molecules.