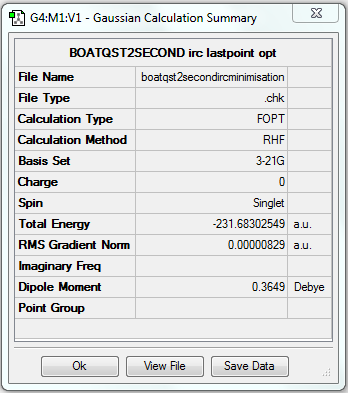
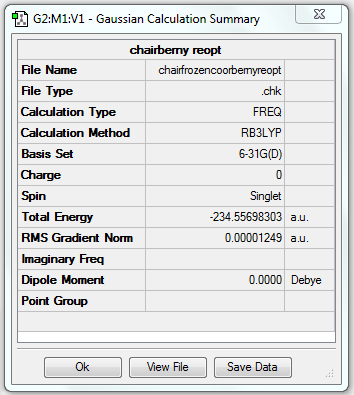
Rep:Mod:yudanchenmod3
The Cope Rearrangement Tutorial
Optimising the Reactants and Products
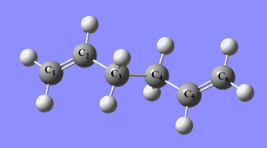
(a) Optimisation of 1,5-hexadiene with an "anti" central linkage
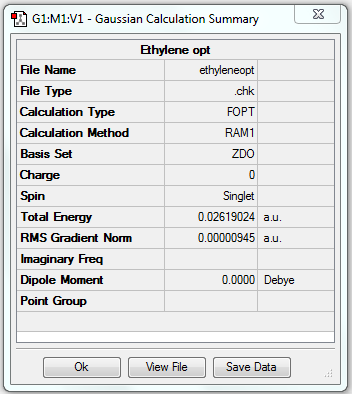
A 1,5-hexadiene molecule was drawn on GaussView by combination of a ethyl fragment and two vinyl fragments. After modifying the dihedral angles of the four C atoms of each end, the anti central linkage was obtained and then optimised.
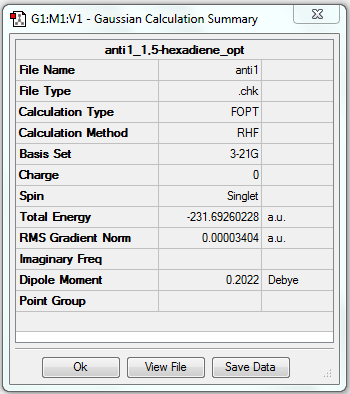
1. Optimising 1,5-hexadiene (anti) using HF/3-21G
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000119 0.000450 YES
RMS Force 0.000018 0.000300 YES
Maximum Displacement 0.001048 0.001800 YES
RMS Displacement 0.000350 0.001200 YES
Predicted change in Energy=-9.131560D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5522 -DE/DX = 0.0001 !
! R2 R(1,3) 1.0869 -DE/DX = 0.0 !
! R3 R(1,4) 1.0836 -DE/DX = 0.0 !
! R4 R(1,7) 1.5089 -DE/DX = 0.0 !
! R5 R(2,5) 1.0869 -DE/DX = 0.0 !
! R6 R(2,6) 1.0836 -DE/DX = 0.0 !
! R7 R(2,12) 1.5089 -DE/DX = 0.0 !
! R8 R(7,8) 1.3161 -DE/DX = 0.0 !
! R9 R(7,9) 1.077 -DE/DX = 0.0 !
! R10 R(8,10) 1.0734 -DE/DX = 0.0 !
! R11 R(8,11) 1.0746 -DE/DX = 0.0 !
! R12 R(12,13) 1.3161 -DE/DX = 0.0 !
! R13 R(12,14) 1.077 -DE/DX = 0.0 !
! R14 R(13,15) 1.0734 -DE/DX = 0.0 !
! R15 R(13,16) 1.0746 -DE/DX = 0.0 !
! A1 A(2,1,3) 108.7763 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.0009 -DE/DX = 0.0 !
! A3 A(2,1,7) 111.376 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.6802 -DE/DX = 0.0 !
! A5 A(3,1,7) 109.614 -DE/DX = 0.0 !
! A6 A(4,1,7) 110.3051 -DE/DX = 0.0 !
! A7 A(1,2,5) 108.7763 -DE/DX = 0.0 !
! A8 A(1,2,6) 109.0009 -DE/DX = 0.0 !
! A9 A(1,2,12) 111.376 -DE/DX = 0.0 !
! A10 A(5,2,6) 107.6802 -DE/DX = 0.0 !
! A11 A(5,2,12) 109.614 -DE/DX = 0.0 !
! A12 A(6,2,12) 110.3051 -DE/DX = 0.0 !
! A13 A(1,7,8) 124.7582 -DE/DX = 0.0 !
! A14 A(1,7,9) 115.539 -DE/DX = 0.0 !
! A15 A(8,7,9) 119.6945 -DE/DX = 0.0 !
! A16 A(7,8,10) 121.8588 -DE/DX = 0.0 !
! A17 A(7,8,11) 121.8063 -DE/DX = 0.0 !
! A18 A(10,8,11) 116.3346 -DE/DX = 0.0 !
! A19 A(2,12,13) 124.7582 -DE/DX = 0.0 !
! A20 A(2,12,14) 115.539 -DE/DX = 0.0 !
! A21 A(13,12,14) 119.6945 -DE/DX = 0.0 !
! A22 A(12,13,15) 121.8588 -DE/DX = 0.0 !
! A23 A(12,13,16) 121.8063 -DE/DX = 0.0 !
! A24 A(15,13,16) 116.3346 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 64.905 -DE/DX = 0.0 !
! D2 D(3,1,2,6) -177.9448 -DE/DX = 0.0 !
! D3 D(3,1,2,12) -56.0129 -DE/DX = 0.0 !
! D4 D(4,1,2,5) -177.9448 -DE/DX = 0.0 !
! D5 D(4,1,2,6) -60.7946 -DE/DX = 0.0 !
! D6 D(4,1,2,12) 61.1373 -DE/DX = 0.0 !
! D7 D(7,1,2,5) -56.0129 -DE/DX = 0.0 !
! D8 D(7,1,2,6) 61.1373 -DE/DX = 0.0 !
! D9 D(7,1,2,12) -176.9308 -DE/DX = 0.0 !
! D10 D(2,1,7,8) -115.1302 -DE/DX = 0.0 !
! D11 D(2,1,7,9) 63.8155 -DE/DX = 0.0 !
! D12 D(3,1,7,8) 124.4436 -DE/DX = 0.0 !
! D13 D(3,1,7,9) -56.6106 -DE/DX = 0.0 !
! D14 D(4,1,7,8) 6.0432 -DE/DX = 0.0 !
! D15 D(4,1,7,9) -175.0111 -DE/DX = 0.0 !
! D16 D(1,2,12,13) -115.1302 -DE/DX = 0.0 !
! D17 D(1,2,12,14) 63.8155 -DE/DX = 0.0 !
! D18 D(5,2,12,13) 124.4436 -DE/DX = 0.0 !
! D19 D(5,2,12,14) -56.6106 -DE/DX = 0.0 !
! D20 D(6,2,12,13) 6.0432 -DE/DX = 0.0 !
! D21 D(6,2,12,14) -175.0111 -DE/DX = 0.0 !
! D22 D(1,7,8,10) 179.1648 -DE/DX = 0.0 !
! D23 D(1,7,8,11) -1.0311 -DE/DX = 0.0 !
! D24 D(9,7,8,10) 0.2598 -DE/DX = 0.0 !
! D25 D(9,7,8,11) -179.9361 -DE/DX = 0.0 !
! D26 D(2,12,13,15) 179.1648 -DE/DX = 0.0 !
! D27 D(2,12,13,16) -1.0311 -DE/DX = 0.0 !
! D28 D(14,12,13,15) 0.2598 -DE/DX = 0.0 !
! D29 D(14,12,13,16) -179.9361 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Linkage | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Anti | Optimisation to a minimum | HF | 3-21G | 250 MB | -231.69260228 a.u. | C2 |
(b) Optimisation of 1,5-hexadiene with an "gauche" central linkage
This molecule was drawn using a similar method as in (a), but the dihedral angles were different. I expect this structure to have higher energy than the one with anti linkage in (a), because for gauche linkage the two large vinyl substituents are closer to each other and thus more sterically hindered, leading to higher energy.
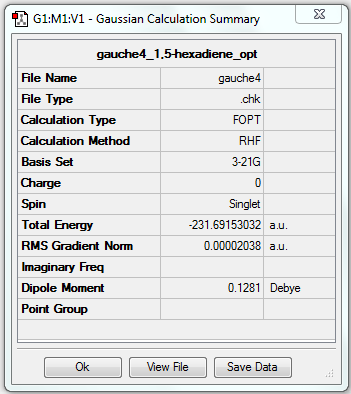
1. Optimising 1,5-hexadiene (gauche) using HF/3-21G
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000078 0.000450 YES
RMS Force 0.000013 0.000300 YES
Maximum Displacement 0.000823 0.001800 YES
RMS Displacement 0.000324 0.001200 YES
Predicted change in Energy=-3.041464D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5555 -DE/DX = -0.0001 !
! R2 R(1,3) 1.0852 -DE/DX = 0.0 !
! R3 R(1,4) 1.0846 -DE/DX = 0.0 !
! R4 R(1,7) 1.5098 -DE/DX = 0.0 !
! R5 R(2,5) 1.0852 -DE/DX = 0.0 !
! R6 R(2,6) 1.0846 -DE/DX = 0.0 !
! R7 R(2,12) 1.5098 -DE/DX = 0.0 !
! R8 R(7,8) 1.3164 -DE/DX = 0.0 !
! R9 R(7,9) 1.0757 -DE/DX = 0.0 !
! R10 R(8,10) 1.0734 -DE/DX = 0.0 !
! R11 R(8,11) 1.0748 -DE/DX = 0.0 !
! R12 R(12,13) 1.3164 -DE/DX = 0.0 !
! R13 R(12,14) 1.0757 -DE/DX = 0.0 !
! R14 R(13,15) 1.0734 -DE/DX = 0.0 !
! R15 R(13,16) 1.0748 -DE/DX = 0.0 !
! A1 A(2,1,3) 108.3062 -DE/DX = 0.0 !
! A2 A(2,1,4) 108.7723 -DE/DX = 0.0 !
! A3 A(2,1,7) 112.3823 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.9558 -DE/DX = 0.0 !
! A5 A(3,1,7) 109.8661 -DE/DX = 0.0 !
! A6 A(4,1,7) 109.4489 -DE/DX = 0.0 !
! A7 A(1,2,5) 108.3062 -DE/DX = 0.0 !
! A8 A(1,2,6) 108.7723 -DE/DX = 0.0 !
! A9 A(1,2,12) 112.3823 -DE/DX = 0.0 !
! A10 A(5,2,6) 107.9558 -DE/DX = 0.0 !
! A11 A(5,2,12) 109.8661 -DE/DX = 0.0 !
! A12 A(6,2,12) 109.4489 -DE/DX = 0.0 !
! A13 A(1,7,8) 124.4233 -DE/DX = 0.0 !
! A14 A(1,7,9) 116.0376 -DE/DX = 0.0 !
! A15 A(8,7,9) 119.5369 -DE/DX = 0.0 !
! A16 A(7,8,10) 121.869 -DE/DX = 0.0 !
! A17 A(7,8,11) 121.8494 -DE/DX = 0.0 !
! A18 A(10,8,11) 116.2813 -DE/DX = 0.0 !
! A19 A(2,12,13) 124.4233 -DE/DX = 0.0 !
! A20 A(2,12,14) 116.0376 -DE/DX = 0.0 !
! A21 A(13,12,14) 119.5369 -DE/DX = 0.0 !
! A22 A(12,13,15) 121.869 -DE/DX = 0.0 !
! A23 A(12,13,16) 121.8494 -DE/DX = 0.0 !
! A24 A(15,13,16) 116.2813 -DE/DX = 0.0 !
! D1 D(3,1,2,5) -179.4769 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 63.4318 -DE/DX = 0.0 !
! D3 D(3,1,2,12) -57.9202 -DE/DX = 0.0 !
! D4 D(4,1,2,5) 63.4318 -DE/DX = 0.0 !
! D5 D(4,1,2,6) -53.6595 -DE/DX = 0.0 !
! D6 D(4,1,2,12) -175.0114 -DE/DX = 0.0 !
! D7 D(7,1,2,5) -57.9202 -DE/DX = 0.0 !
! D8 D(7,1,2,6) -175.0114 -DE/DX = 0.0 !
! D9 D(7,1,2,12) 63.6366 -DE/DX = 0.0 !
! D10 D(2,1,7,8) 109.4257 -DE/DX = 0.0 !
! D11 D(2,1,7,9) -70.0344 -DE/DX = 0.0 !
! D12 D(3,1,7,8) -129.912 -DE/DX = 0.0 !
! D13 D(3,1,7,9) 50.6278 -DE/DX = 0.0 !
! D14 D(4,1,7,8) -11.5386 -DE/DX = 0.0 !
! D15 D(4,1,7,9) 169.0013 -DE/DX = 0.0 !
! D16 D(1,2,12,13) 109.4257 -DE/DX = 0.0 !
! D17 D(1,2,12,14) -70.0344 -DE/DX = 0.0 !
! D18 D(5,2,12,13) -129.912 -DE/DX = 0.0 !
! D19 D(5,2,12,14) 50.6278 -DE/DX = 0.0 !
! D20 D(6,2,12,13) -11.5386 -DE/DX = 0.0 !
! D21 D(6,2,12,14) 169.0013 -DE/DX = 0.0 !
! D22 D(1,7,8,10) -179.1965 -DE/DX = 0.0 !
! D23 D(1,7,8,11) 1.0282 -DE/DX = 0.0 !
! D24 D(9,7,8,10) 0.2459 -DE/DX = 0.0 !
! D25 D(9,7,8,11) -179.5294 -DE/DX = 0.0 !
! D26 D(2,12,13,15) -179.1965 -DE/DX = 0.0 !
! D27 D(2,12,13,16) 1.0282 -DE/DX = 0.0 !
! D28 D(14,12,13,15) 0.2459 -DE/DX = 0.0 !
! D29 D(14,12,13,16) -179.5294 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Linkage | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Gauche | Optimisation to a minimum | HF | 3-21G | 250 MB | -231.69153032 a.u. | C2 |
7. Comparison with (a)
| Energy (a) | Energy (b) | Energy difference (b)-(a) |
|---|---|---|
| -231.69260228 a.u. | -231.69153032 a.u. | 0.00107196 a.u. |
Just like that I expected, the final energy of the optimised structure with gauche linkage is higher than that with anti linkage due to the more hindered substituents. The energy difference is equal to 0.00107196 a.u.(or 0.6726656196 kcal/mol).
(c) Optimisation of lowest energy conformation of 1,5-hexadiene
From my perspective, the structures with gauche linkage should have higher energy than those with anti linkage because of the stronger hinderance between vinyl substituents, just like the result obtained from (a) and (b) above. And this is also the general trend for butane. Therefore in order to test my hypothesis, I tried to optimise another structure with gauche linkage to see if the energy is still higher than that obtained in (a).
1. Optimising 1,5-hexadiene (gauche) using HF/3-21G
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000030 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.001272 0.001800 YES
RMS Displacement 0.000469 0.001200 YES
Predicted change in Energy=-1.907270D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5532 -DE/DX = 0.0 !
! R2 R(1,3) 1.0836 -DE/DX = 0.0 !
! R3 R(1,4) 1.0867 -DE/DX = 0.0 !
! R4 R(1,12) 1.5093 -DE/DX = 0.0 !
! R5 R(2,5) 1.0872 -DE/DX = 0.0 !
! R6 R(2,6) 1.0844 -DE/DX = 0.0 !
! R7 R(2,7) 1.5085 -DE/DX = 0.0 !
! R8 R(7,8) 1.3163 -DE/DX = 0.0 !
! R9 R(7,9) 1.0751 -DE/DX = 0.0 !
! R10 R(8,10) 1.0735 -DE/DX = 0.0 !
! R11 R(8,11) 1.0748 -DE/DX = 0.0 !
! R12 R(12,13) 1.3165 -DE/DX = 0.0 !
! R13 R(12,14) 1.0773 -DE/DX = 0.0 !
! R14 R(13,15) 1.0734 -DE/DX = 0.0 !
! R15 R(13,16) 1.0745 -DE/DX = 0.0 !
! A1 A(2,1,3) 109.191 -DE/DX = 0.0 !
! A2 A(2,1,4) 108.4571 -DE/DX = 0.0 !
! A3 A(2,1,12) 111.8663 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.8761 -DE/DX = 0.0 !
! A5 A(3,1,12) 110.2825 -DE/DX = 0.0 !
! A6 A(4,1,12) 109.0666 -DE/DX = 0.0 !
! A7 A(1,2,5) 108.6427 -DE/DX = 0.0 !
! A8 A(1,2,6) 109.3183 -DE/DX = 0.0 !
! A9 A(1,2,7) 111.7764 -DE/DX = 0.0 !
! A10 A(5,2,6) 107.5329 -DE/DX = 0.0 !
! A11 A(5,2,7) 109.7284 -DE/DX = 0.0 !
! A12 A(6,2,7) 109.7401 -DE/DX = 0.0 !
! A13 A(2,7,8) 124.532 -DE/DX = 0.0 !
! A14 A(2,7,9) 115.5484 -DE/DX = 0.0 !
! A15 A(8,7,9) 119.9131 -DE/DX = 0.0 !
! A16 A(7,8,10) 121.7749 -DE/DX = 0.0 !
! A17 A(7,8,11) 121.9613 -DE/DX = 0.0 !
! A18 A(10,8,11) 116.2638 -DE/DX = 0.0 !
! A19 A(1,12,13) 125.0268 -DE/DX = 0.0 !
! A20 A(1,12,14) 115.3004 -DE/DX = 0.0 !
! A21 A(13,12,14) 119.6721 -DE/DX = 0.0 !
! A22 A(12,13,15) 121.843 -DE/DX = 0.0 !
! A23 A(12,13,16) 121.7798 -DE/DX = 0.0 !
! A24 A(15,13,16) 116.3769 -DE/DX = 0.0 !
! D1 D(3,1,2,5) -175.8704 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 67.0538 -DE/DX = 0.0 !
! D3 D(3,1,2,7) -54.645 -DE/DX = 0.0 !
! D4 D(4,1,2,5) 66.8188 -DE/DX = 0.0 !
! D5 D(4,1,2,6) -50.257 -DE/DX = 0.0 !
! D6 D(4,1,2,7) -171.9558 -DE/DX = 0.0 !
! D7 D(12,1,2,5) -53.5141 -DE/DX = 0.0 !
! D8 D(12,1,2,6) -170.5899 -DE/DX = 0.0 !
! D9 D(12,1,2,7) 67.7114 -DE/DX = 0.0 !
! D10 D(2,1,12,13) -117.2463 -DE/DX = 0.0 !
! D11 D(2,1,12,14) 62.4359 -DE/DX = 0.0 !
! D12 D(3,1,12,13) 4.4843 -DE/DX = 0.0 !
! D13 D(3,1,12,14) -175.8335 -DE/DX = 0.0 !
! D14 D(4,1,12,13) 122.7772 -DE/DX = 0.0 !
! D15 D(4,1,12,14) -57.5406 -DE/DX = 0.0 !
! D16 D(1,2,7,8) 120.8433 -DE/DX = 0.0 !
! D17 D(1,2,7,9) -58.2268 -DE/DX = 0.0 !
! D18 D(5,2,7,8) -118.5622 -DE/DX = 0.0 !
! D19 D(5,2,7,9) 62.3677 -DE/DX = 0.0 !
! D20 D(6,2,7,8) -0.6121 -DE/DX = 0.0 !
! D21 D(6,2,7,9) -179.6822 -DE/DX = 0.0 !
! D22 D(2,7,8,10) -179.4496 -DE/DX = 0.0 !
! D23 D(2,7,8,11) 0.6474 -DE/DX = 0.0 !
! D24 D(9,7,8,10) -0.4176 -DE/DX = 0.0 !
! D25 D(9,7,8,11) 179.6795 -DE/DX = 0.0 !
! D26 D(1,12,13,15) 179.8587 -DE/DX = 0.0 !
! D27 D(1,12,13,16) -0.324 -DE/DX = 0.0 !
! D28 D(14,12,13,15) 0.1893 -DE/DX = 0.0 !
! D29 D(14,12,13,16) -179.9933 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Linkage | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
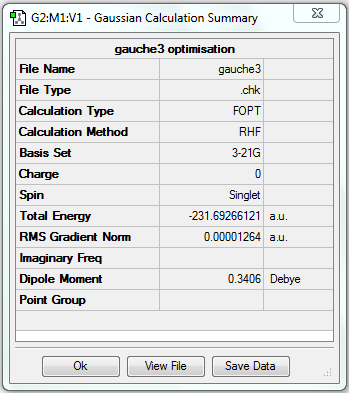
| Gauche | Optimisation to a minimum | HF | 3-21G | Default | -231.69266121 a.u. | C1 |
7. Comparison with (a)
| Energy (a) | Energy (c) | Energy difference (a)-(c) |
|---|---|---|
| -231.69260228 a.u. | -231.69266121 a.u. | 0.00005893 a.u. |
Surprisingly in this case, the final energy of the optimised structure with gauche linkage is lower than that with anti linkage, which is opposite to my hypothesis. The energy difference is equal to 0.00005893 a.u.(or 0.0369791643 kcal/mol). Perhaps this is because for 1,5-hexadiene, there is another factor affecting the stability of conformers, that is, the π bonds of neighbouring vinyl groups may have π/π* interactions leading to extra stablisations, so there may be some particular structures with gauche linkage having lower energy than those with anti linkage. As the structure obtained in (c) has a lower energy than that in either (a) or (b), it is likely to be the lowest energy conformer of 1,5-hexadiene.
(d) Identification of optimised structures
| Optimised stucture | Conformer identified from Appendix 1 |
|---|---|
| (a) | Anti1 |
| (b) | Gauche4 |
| (c) | Gauche3 |
(e) Optimisation of anti2 conformer using HF/3-21G
1. Optimising 1,5-hexadiene (anti2) using HF/3-21G
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000121 0.000450 YES
RMS Force 0.000018 0.000300 YES
Maximum Displacement 0.000593 0.001800 YES
RMS Displacement 0.000192 0.001200 YES
Predicted change in Energy=-1.010799D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5526 -DE/DX = 0.0001 !
! R2 R(1,3) 1.0856 -DE/DX = 0.0 !
! R3 R(1,4) 1.0848 -DE/DX = 0.0 !
! R4 R(1,7) 1.509 -DE/DX = 0.0 !
! R5 R(2,5) 1.0856 -DE/DX = 0.0 !
! R6 R(2,6) 1.0848 -DE/DX = 0.0 !
! R7 R(2,12) 1.509 -DE/DX = 0.0 !
! R8 R(7,8) 1.3161 -DE/DX = 0.0 !
! R9 R(7,9) 1.0769 -DE/DX = 0.0 !
! R10 R(8,10) 1.0734 -DE/DX = 0.0 !
! R11 R(8,11) 1.0746 -DE/DX = 0.0 !
! R12 R(12,13) 1.3161 -DE/DX = 0.0 !
! R13 R(12,14) 1.0769 -DE/DX = 0.0 !
! R14 R(13,15) 1.0734 -DE/DX = 0.0 !
! R15 R(13,16) 1.0746 -DE/DX = 0.0 !
! A1 A(2,1,3) 108.3477 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4093 -DE/DX = 0.0 !
! A3 A(2,1,7) 111.351 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.7104 -DE/DX = 0.0 !
! A5 A(3,1,7) 109.972 -DE/DX = 0.0 !
! A6 A(4,1,7) 109.9636 -DE/DX = 0.0 !
! A7 A(1,2,5) 108.3477 -DE/DX = 0.0 !
! A8 A(1,2,6) 109.4093 -DE/DX = 0.0 !
! A9 A(1,2,12) 111.351 -DE/DX = 0.0 !
! A10 A(5,2,6) 107.7104 -DE/DX = 0.0 !
! A11 A(5,2,12) 109.972 -DE/DX = 0.0 !
! A12 A(6,2,12) 109.9636 -DE/DX = 0.0 !
! A13 A(1,7,8) 124.8104 -DE/DX = 0.0 !
! A14 A(1,7,9) 115.5094 -DE/DX = 0.0 !
! A15 A(8,7,9) 119.6718 -DE/DX = 0.0 !
! A16 A(7,8,10) 121.8643 -DE/DX = 0.0 !
! A17 A(7,8,11) 121.822 -DE/DX = 0.0 !
! A18 A(10,8,11) 116.3134 -DE/DX = 0.0 !
! A19 A(2,12,13) 124.8104 -DE/DX = 0.0 !
! A20 A(2,12,14) 115.5094 -DE/DX = 0.0 !
! A21 A(13,12,14) 119.6718 -DE/DX = 0.0 !
! A22 A(12,13,15) 121.8643 -DE/DX = 0.0 !
! A23 A(12,13,16) 121.822 -DE/DX = 0.0 !
! A24 A(15,13,16) 116.3134 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 180.0 -DE/DX = 0.0 !
! D2 D(3,1,2,6) -62.8281 -DE/DX = 0.0 !
! D3 D(3,1,2,12) 58.9347 -DE/DX = 0.0 !
! D4 D(4,1,2,5) 62.8281 -DE/DX = 0.0 !
! D5 D(4,1,2,6) 180.0 -DE/DX = 0.0 !
! D6 D(4,1,2,12) -58.2372 -DE/DX = 0.0 !
! D7 D(7,1,2,5) -58.9347 -DE/DX = 0.0 !
! D8 D(7,1,2,6) 58.2372 -DE/DX = 0.0 !
! D9 D(7,1,2,12) 180.0 -DE/DX = 0.0 !
! D10 D(2,1,7,8) 114.6503 -DE/DX = 0.0 !
! D11 D(2,1,7,9) -64.2825 -DE/DX = 0.0 !
! D12 D(3,1,7,8) -125.2392 -DE/DX = 0.0 !
! D13 D(3,1,7,9) 55.828 -DE/DX = 0.0 !
! D14 D(4,1,7,8) -6.7902 -DE/DX = 0.0 !
! D15 D(4,1,7,9) 174.277 -DE/DX = 0.0 !
! D16 D(1,2,12,13) -114.6503 -DE/DX = 0.0 !
! D17 D(1,2,12,14) 64.2825 -DE/DX = 0.0 !
! D18 D(5,2,12,13) 125.2392 -DE/DX = 0.0 !
! D19 D(5,2,12,14) -55.828 -DE/DX = 0.0 !
! D20 D(6,2,12,13) 6.7902 -DE/DX = 0.0 !
! D21 D(6,2,12,14) -174.277 -DE/DX = 0.0 !
! D22 D(1,7,8,10) -179.126 -DE/DX = 0.0 !
! D23 D(1,7,8,11) 1.1023 -DE/DX = 0.0 !
! D24 D(9,7,8,10) -0.2346 -DE/DX = 0.0 !
! D25 D(9,7,8,11) -180.0063 -DE/DX = 0.0 !
! D26 D(2,12,13,15) 179.126 -DE/DX = 0.0 !
! D27 D(2,12,13,16) -1.1023 -DE/DX = 0.0 !
! D28 D(14,12,13,15) 0.2346 -DE/DX = 0.0 !
! D29 D(14,12,13,16) 180.0063 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Conformer | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
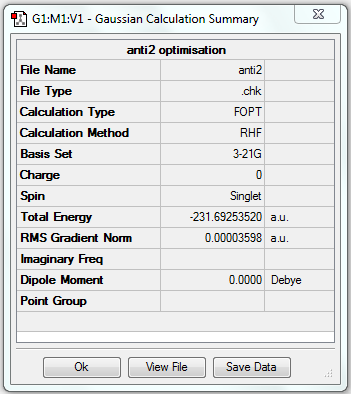
| Anti2 | Optimisation to a minimum | HF | 3-21G | Default | -231.69253520 a.u. | Ci |
7. Comparison with Appendix 1
| Energy (optimised) | Energy (Appendix 1) |
|---|---|
| -231.69253520 a.u. | -231.69254 a.u. |
The energy is very similar for my optimised structure and the one in Appendix 1, confirming the structures are the same.
(f) Reoptimisation of anti2 conformer using B3LYP/6-31G(d)
In this section, I reoptimised the previously HF/3-21G optimised anti2 1,5-hexadiene using a better method and basis set i.e. B3LYP/6-31G(d) in order to obtain a more accurate energy minimum.
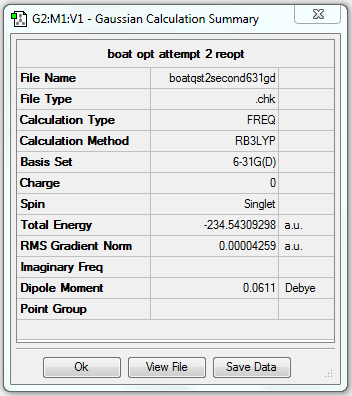
1. Optimising 1,5-hexadiene (anti2) using B3LYP/6-31G(d)
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000015 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000226 0.001800 YES
RMS Displacement 0.000081 0.001200 YES
Predicted change in Energy=-1.602862D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5481 -DE/DX = 0.0 !
! R2 R(1,3) 1.0997 -DE/DX = 0.0 !
! R3 R(1,4) 1.098 -DE/DX = 0.0 !
! R4 R(1,7) 1.5042 -DE/DX = 0.0 !
! R5 R(2,5) 1.0997 -DE/DX = 0.0 !
! R6 R(2,6) 1.098 -DE/DX = 0.0 !
! R7 R(2,12) 1.5042 -DE/DX = 0.0 !
! R8 R(7,8) 1.3335 -DE/DX = 0.0 !
! R9 R(7,9) 1.0919 -DE/DX = 0.0 !
! R10 R(8,10) 1.0868 -DE/DX = 0.0 !
! R11 R(8,11) 1.0885 -DE/DX = 0.0 !
! R12 R(12,13) 1.3335 -DE/DX = 0.0 !
! R13 R(12,14) 1.0919 -DE/DX = 0.0 !
! R14 R(13,15) 1.0868 -DE/DX = 0.0 !
! R15 R(13,16) 1.0885 -DE/DX = 0.0 !
! A1 A(2,1,3) 108.1899 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.6058 -DE/DX = 0.0 !
! A3 A(2,1,7) 112.6721 -DE/DX = 0.0 !
! A4 A(3,1,4) 106.66 -DE/DX = 0.0 !
! A5 A(3,1,7) 109.7814 -DE/DX = 0.0 !
! A6 A(4,1,7) 109.742 -DE/DX = 0.0 !
! A7 A(1,2,5) 108.1899 -DE/DX = 0.0 !
! A8 A(1,2,6) 109.6058 -DE/DX = 0.0 !
! A9 A(1,2,12) 112.6721 -DE/DX = 0.0 !
! A10 A(5,2,6) 106.66 -DE/DX = 0.0 !
! A11 A(5,2,12) 109.7814 -DE/DX = 0.0 !
! A12 A(6,2,12) 109.742 -DE/DX = 0.0 !
! A13 A(1,7,8) 125.2867 -DE/DX = 0.0 !
! A14 A(1,7,9) 115.7272 -DE/DX = 0.0 !
! A15 A(8,7,9) 118.9814 -DE/DX = 0.0 !
! A16 A(7,8,10) 121.8701 -DE/DX = 0.0 !
! A17 A(7,8,11) 121.6516 -DE/DX = 0.0 !
! A18 A(10,8,11) 116.4778 -DE/DX = 0.0 !
! A19 A(2,12,13) 125.2867 -DE/DX = 0.0 !
! A20 A(2,12,14) 115.7272 -DE/DX = 0.0 !
! A21 A(13,12,14) 118.9814 -DE/DX = 0.0 !
! A22 A(12,13,15) 121.8701 -DE/DX = 0.0 !
! A23 A(12,13,16) 121.6516 -DE/DX = 0.0 !
! A24 A(15,13,16) 116.4778 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 180.0 -DE/DX = 0.0 !
! D2 D(3,1,2,6) -64.0627 -DE/DX = 0.0 !
! D3 D(3,1,2,12) 58.444 -DE/DX = 0.0 !
! D4 D(4,1,2,5) 64.0627 -DE/DX = 0.0 !
! D5 D(4,1,2,6) 180.0 -DE/DX = 0.0 !
! D6 D(4,1,2,12) -57.4932 -DE/DX = 0.0 !
! D7 D(7,1,2,5) -58.444 -DE/DX = 0.0 !
! D8 D(7,1,2,6) 57.4932 -DE/DX = 0.0 !
! D9 D(7,1,2,12) -180.0 -DE/DX = 0.0 !
! D10 D(2,1,7,8) 118.5282 -DE/DX = 0.0 !
! D11 D(2,1,7,9) -60.6754 -DE/DX = 0.0 !
! D12 D(3,1,7,8) -120.8235 -DE/DX = 0.0 !
! D13 D(3,1,7,9) 59.9729 -DE/DX = 0.0 !
! D14 D(4,1,7,8) -3.9021 -DE/DX = 0.0 !
! D15 D(4,1,7,9) 176.8943 -DE/DX = 0.0 !
! D16 D(1,2,12,13) -118.5282 -DE/DX = 0.0 !
! D17 D(1,2,12,14) 60.6754 -DE/DX = 0.0 !
! D18 D(5,2,12,13) 120.8235 -DE/DX = 0.0 !
! D19 D(5,2,12,14) -59.9729 -DE/DX = 0.0 !
! D20 D(6,2,12,13) 3.9021 -DE/DX = 0.0 !
! D21 D(6,2,12,14) -176.8943 -DE/DX = 0.0 !
! D22 D(1,7,8,10) -179.5641 -DE/DX = 0.0 !
! D23 D(1,7,8,11) 0.7139 -DE/DX = 0.0 !
! D24 D(9,7,8,10) -0.3843 -DE/DX = 0.0 !
! D25 D(9,7,8,11) 179.8938 -DE/DX = 0.0 !
! D26 D(2,12,13,15) 179.5641 -DE/DX = 0.0 !
! D27 D(2,12,13,16) -0.7139 -DE/DX = 0.0 !
! D28 D(14,12,13,15) 0.3843 -DE/DX = 0.0 !
! D29 D(14,12,13,16) -179.8938 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Conformer | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
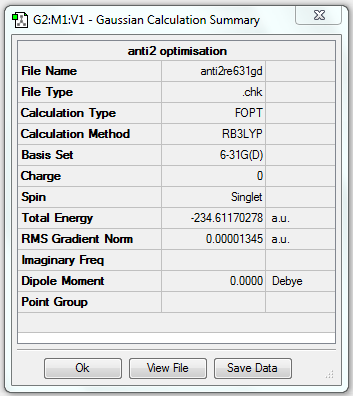
| Anti2 | Optimisation to a minimum | B3LYP | 6-31G(d) | Default | -234.61170278 a.u. | Ci |
7. Comparison with (e)
| Energy (HF/3-21G) | Energy (B3LYP/6-31G(d)) | Energy difference |
|---|---|---|
| -231.69253520 a.u. | -234.61170278 a.u. | 2.91916758 a.u. |
The energy of B3LYP/6-31G(d) optimised structure is much lower than that of HF/3-21G optimised structure, and the energy difference is equal to 2.91916758 a.u.(or 1831.8068481258 kcal/mol). However, there are no visible differences between the two structures by simply looking at them on GaussView as the following is shown.
| HF/3-21G | B3LYP/6-31G(d) | |||||||
|---|---|---|---|---|---|---|---|---|
| Structure |
|
|
Therefore we need to compare the geometric data between the two structures as the table is shown below, so as to find out the change in geometry responsible for the large energy difference.
| Geometric parameter | HF/3-21G | B3LYP/6-31G(d) |
|---|---|---|
| C1=C2 (or C5=C6) bond length | 1.31614 Å | 1.33352 Å |
| C2-C3 (or C4=C5) bond length | 1.50895 Å | 1.50419 Å |
| C3-C4 bond length | 1.55262 Å | 1.54814 Å |
| C1=C2-C3-C4 (or C3-C4-C5=C6) dihedral angle | +(or-)114.65024o | +(or-)118.52823o |
Overall, the geometry change is small, but the change in dihedral angles may be the main reason for the large energy difference. Perhaps for the B3LYP/6-31G(d) optimised structure, the double bonds have a better alignment with the neighbouring C-C/C-H bonds, resulting in strong σ-π conjugations and thus have a large stablisation in energy.
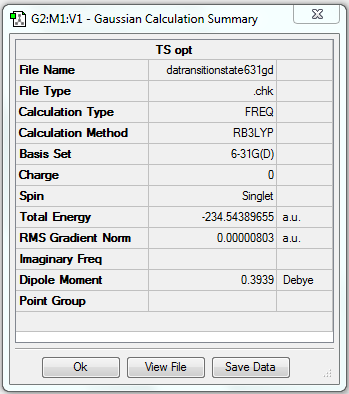
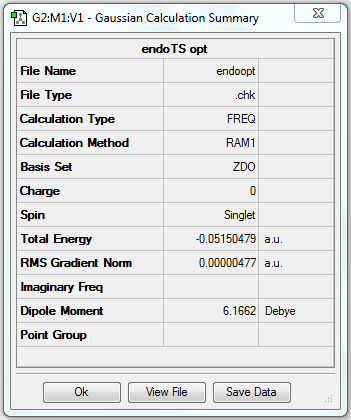
(g) Frequency analysis of optimised anti2 structure
The frequency analysis is the second derivative of the potential energy surface of a reaction. By looking at the vibrational frequency table, we can determine whether the former optimisation is successful or not. And it provides useful information of IR active and Raman active vibrations.
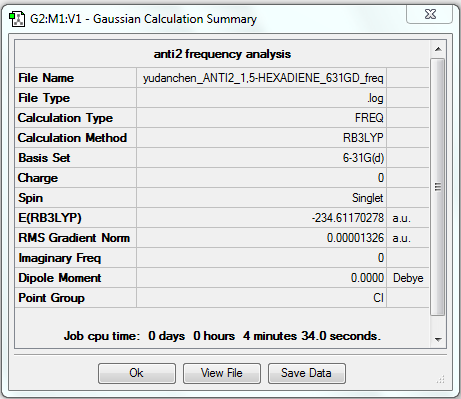
Frequency analysis of B3LYP/6-31G(d) optimised anti2 structure
1. File link
The frequency analysed file is linked to here.
The energy is the same as that obtained in optimisation, which means the structure is correct (i.e. the same as the optimised structure).
3. Real output
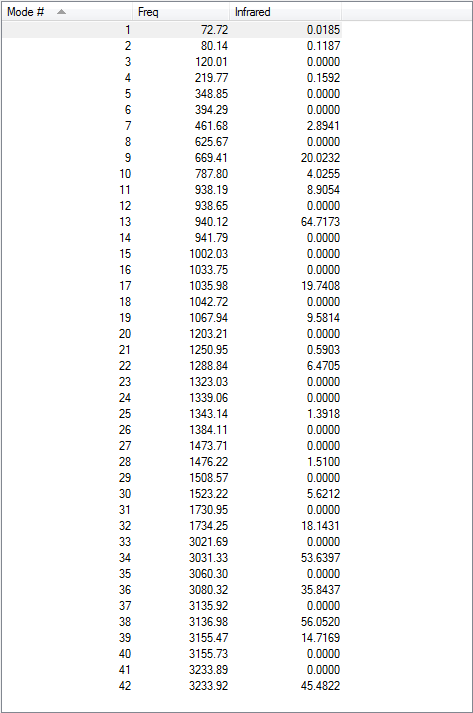
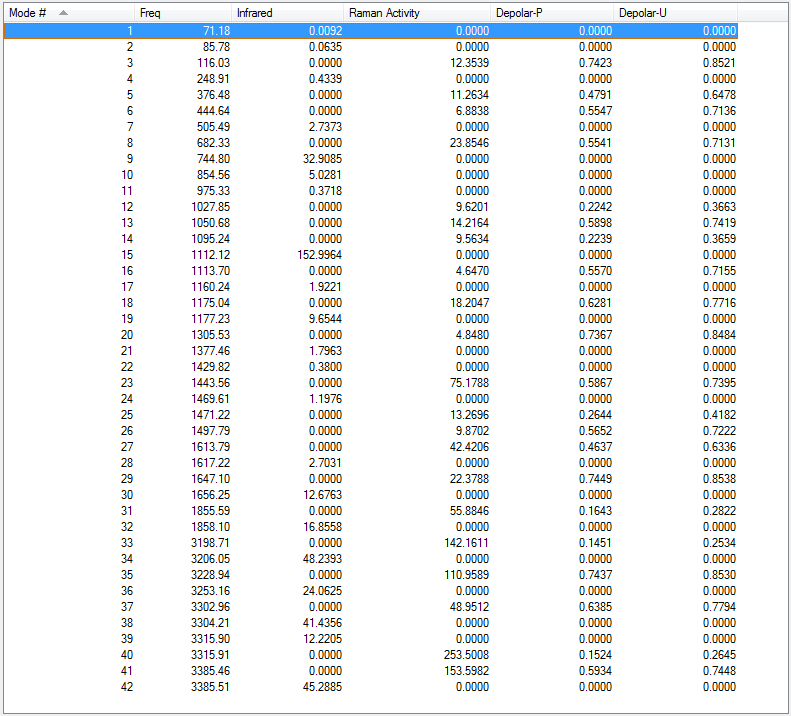
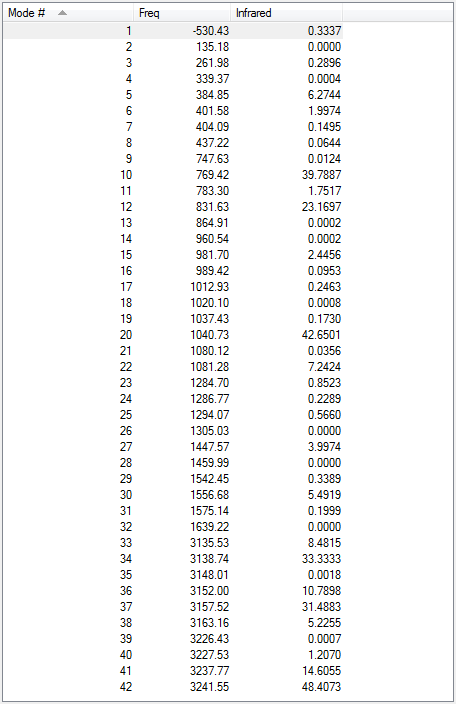
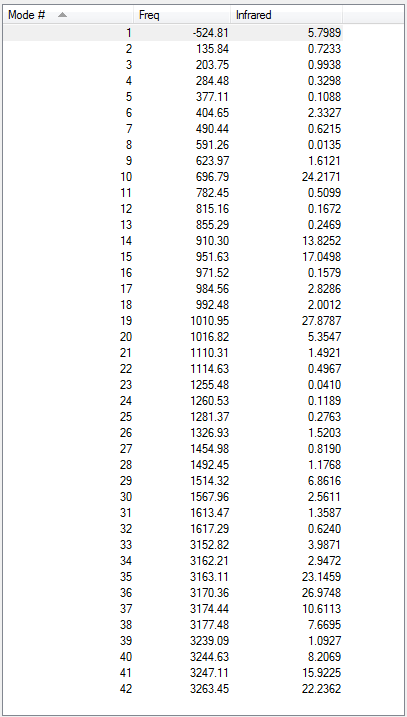
Low frequencies --- -18.7010 -11.7206 0.0009 0.0009 0.0011 1.7692 Low frequencies --- 72.7203 80.1406 120.0180
All vibrational frequencies are real and positive, indicating the molecule is fully optimised.
The vibrational frequency table gives many vibrations that have zero IR absorption intensity and thus do not show on the IR spectrum, which is because the high symmetry of anti2 1,5-hexadiene i.e. Ci, leading to lack of dipole moment on some particular vibrations that are IR inactive.
6. Thermochemistry
Sum of electronic and zero-point Energies= -234.469212 Sum of electronic and thermal Energies= -234.461856 Sum of electronic and thermal Enthalpies= -234.460912 Sum of electronic and thermal Free Energies= -234.500821
7. Key information
| Conformer | Job type | Method | Basis set | Memory limit | Energy |
|---|---|---|---|---|---|
| Anti2 | Frequency | B3LYP | 6-31G(d) | Default | -234.61170278 a.u. |
Frequency analysis of HF/3-21G optimised anti2 structure
1. File link
The frequency analysed file is linked to here.
The energy is the same as that obtained in optimisation, which means the structure is correct (i.e. the same as the optimised structure).
3. Real output
Low frequencies --- -7.4353 -2.0270 -0.0015 -0.0014 -0.0014 2.0047 Low frequencies --- 71.1769 85.7840 116.0337
The low frequencies are within ±15 cm-1.
All vibrational frequencies are real and positive, indicating the molecule is fully optimised.
The vibrational frequency table gives many vibrations that have zero IR absorption intensity and thus do not show on the IR spectrum, which is because the high symmetry of anti2 1,5-hexadiene i.e. Ci, leading to lack of dipole moment on some particular vibrations that are IR inactive.
6. Thermochemistry
Sum of electronic and zero-point Energies= -231.539539 Sum of electronic and thermal Energies= -231.532565 Sum of electronic and thermal Enthalpies= -231.531621 Sum of electronic and thermal Free Energies= -231.570915
7. Key information
| Conformer | Job type | Method | Basis set | Memory limit | Energy |
|---|---|---|---|---|---|
| Anti2 | Frequency | HF | 3-21G | Default | -231.69253520 a.u. |
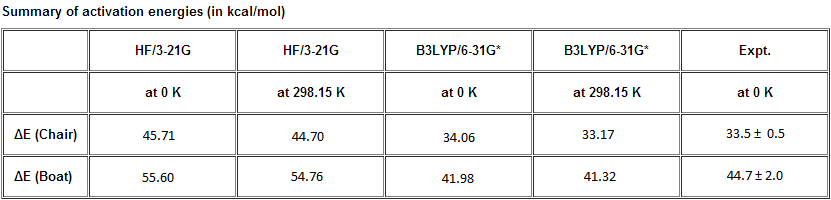
Optimising the "Chair" and "Boat" Transition Structures
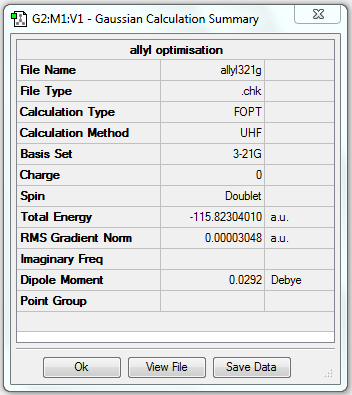
(a) Optimisation of allyl fragment
In order to build up the transition state, an optimised allyl fragment is needed, which can combine with another one in two different orientations to give the chair or boat structure.
1. Optimising allyl fragment using HF/3-21G
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000048 0.000450 YES
RMS Force 0.000018 0.000300 YES
Maximum Displacement 0.000149 0.001800 YES
RMS Displacement 0.000070 0.001200 YES
Predicted change in Energy=-1.277266D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3885 -DE/DX = 0.0 !
! R2 R(1,3) 1.0722 -DE/DX = 0.0 !
! R3 R(1,4) 1.074 -DE/DX = 0.0 !
! R4 R(2,5) 1.0756 -DE/DX = 0.0 !
! R5 R(2,6) 1.3885 -DE/DX = 0.0 !
! R6 R(6,7) 1.074 -DE/DX = 0.0 !
! R7 R(6,8) 1.0722 -DE/DX = 0.0 !
! A1 A(2,1,3) 121.4197 -DE/DX = 0.0 !
! A2 A(2,1,4) 121.1212 -DE/DX = 0.0 !
! A3 A(3,1,4) 117.4591 -DE/DX = 0.0 !
! A4 A(1,2,5) 117.8473 -DE/DX = 0.0 !
! A5 A(1,2,6) 124.3054 -DE/DX = 0.0 !
! A6 A(5,2,6) 117.8473 -DE/DX = 0.0 !
! A7 A(2,6,7) 121.1212 -DE/DX = 0.0 !
! A8 A(2,6,8) 121.4197 -DE/DX = 0.0 !
! A9 A(7,6,8) 117.4591 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 0.0 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 180.0 -DE/DX = 0.0 !
! D3 D(4,1,2,5) 180.0 -DE/DX = 0.0 !
! D4 D(4,1,2,6) 0.0 -DE/DX = 0.0 !
! D5 D(1,2,6,7) 0.0 -DE/DX = 0.0 !
! D6 D(1,2,6,8) 180.0 -DE/DX = 0.0 !
! D7 D(5,2,6,7) 180.0 -DE/DX = 0.0 !
! D8 D(5,2,6,8) 0.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Fragment | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Allyl | Optimisation to a minimum | HF | 3-21G | Default | -115.82304010 a.u. | C2v |
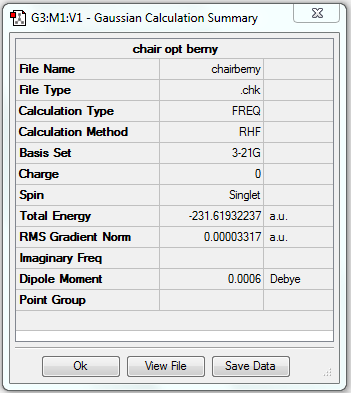
(b) Optimisation of chair transition state by computing force constants
This optimisation method is very easy and quick, but it only works well for any guess transition state that is already close enough to the exact structure.
1. Optimising chair transition state using Berny method with force constants calculation
test molecule |
The optimised structure looks very similar to the one in Appendix 2 on the right.
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000059 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.001059 0.001800 YES
RMS Displacement 0.000295 0.001200 YES
Predicted change in Energy=-1.719612D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3893 -DE/DX = 0.0 !
! R2 R(1,3) 1.076 -DE/DX = 0.0 !
! R3 R(1,4) 1.0743 -DE/DX = 0.0 !
! R4 R(1,12) 2.3921 -DE/DX = 0.0 !
! R5 R(2,5) 1.0759 -DE/DX = 0.0 !
! R6 R(2,6) 1.3894 -DE/DX = -0.0001 !
! R7 R(4,9) 2.3918 -DE/DX = 0.0 !
! R8 R(6,7) 1.0742 -DE/DX = 0.0 !
! R9 R(6,8) 1.076 -DE/DX = 0.0 !
! R10 R(6,15) 2.3921 -DE/DX = 0.0 !
! R11 R(7,14) 2.3921 -DE/DX = 0.0 !
! R12 R(9,10) 1.3892 -DE/DX = 0.0 !
! R13 R(9,11) 1.076 -DE/DX = 0.0 !
! R14 R(9,12) 1.0741 -DE/DX = 0.0001 !
! R15 R(10,13) 1.0759 -DE/DX = 0.0 !
! R16 R(10,14) 1.3893 -DE/DX = 0.0 !
! R17 R(14,15) 1.0743 -DE/DX = 0.0 !
! R18 R(14,16) 1.076 -DE/DX = 0.0 !
! A1 A(2,1,3) 119.0177 -DE/DX = 0.0 !
! A2 A(2,1,4) 118.8781 -DE/DX = 0.0 !
! A3 A(2,1,12) 90.503 -DE/DX = 0.0 !
! A4 A(3,1,4) 113.8221 -DE/DX = 0.0 !
! A5 A(3,1,12) 85.538 -DE/DX = 0.0 !
! A6 A(4,1,12) 122.638 -DE/DX = 0.0 !
! A7 A(1,2,5) 118.198 -DE/DX = 0.0 !
! A8 A(1,2,6) 120.4933 -DE/DX = 0.0 !
! A9 A(5,2,6) 118.193 -DE/DX = 0.0 !
! A10 A(1,4,9) 57.0801 -DE/DX = 0.0 !
! A11 A(2,6,7) 118.8686 -DE/DX = 0.0 !
! A12 A(2,6,8) 119.015 -DE/DX = 0.0 !
! A13 A(2,6,15) 90.4821 -DE/DX = 0.0 !
! A14 A(7,6,8) 113.8145 -DE/DX = 0.0 !
! A15 A(7,6,15) 122.6774 -DE/DX = 0.0 !
! A16 A(8,6,15) 85.5566 -DE/DX = 0.0 !
! A17 A(6,7,14) 57.056 -DE/DX = 0.0 !
! A18 A(4,9,10) 90.5059 -DE/DX = 0.0 !
! A19 A(4,9,11) 85.5501 -DE/DX = 0.0 !
! A20 A(4,9,12) 122.6678 -DE/DX = 0.0 !
! A21 A(10,9,11) 119.019 -DE/DX = 0.0 !
! A22 A(10,9,12) 118.8612 -DE/DX = 0.0 !
! A23 A(11,9,12) 113.8138 -DE/DX = 0.0 !
! A24 A(9,10,13) 118.1924 -DE/DX = 0.0 !
! A25 A(9,10,14) 120.5014 -DE/DX = 0.0 !
! A26 A(13,10,14) 118.1916 -DE/DX = 0.0 !
! A27 A(1,12,9) 57.0668 -DE/DX = 0.0 !
! A28 A(7,14,10) 90.4787 -DE/DX = 0.0 !
! A29 A(7,14,15) 122.668 -DE/DX = 0.0 !
! A30 A(7,14,16) 85.5339 -DE/DX = 0.0 !
! A31 A(10,14,15) 118.86 -DE/DX = 0.0 !
! A32 A(10,14,16) 119.0207 -DE/DX = 0.0 !
! A33 A(15,14,16) 113.8381 -DE/DX = 0.0 !
! A34 A(6,15,14) 57.0579 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 18.0655 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 177.7575 -DE/DX = 0.0 !
! D3 D(4,1,2,5) 164.5162 -DE/DX = 0.0 !
! D4 D(4,1,2,6) -35.7917 -DE/DX = 0.0 !
! D5 D(12,1,2,5) -67.1103 -DE/DX = 0.0 !
! D6 D(12,1,2,6) 92.5817 -DE/DX = 0.0 !
! D7 D(2,1,4,9) 107.3365 -DE/DX = 0.0 !
! D8 D(3,1,4,9) -104.5532 -DE/DX = 0.0 !
! D9 D(12,1,4,9) -4.0796 -DE/DX = 0.0 !
! D10 D(2,1,12,9) -116.2747 -DE/DX = 0.0 !
! D11 D(3,1,12,9) 124.6546 -DE/DX = 0.0 !
! D12 D(4,1,12,9) 9.1162 -DE/DX = 0.0 !
! D13 D(1,2,6,7) 35.8021 -DE/DX = 0.0 !
! D14 D(1,2,6,8) -177.7862 -DE/DX = 0.0 !
! D15 D(1,2,6,15) -92.6007 -DE/DX = 0.0 !
! D16 D(5,2,6,7) -164.5049 -DE/DX = 0.0 !
! D17 D(5,2,6,8) -18.0931 -DE/DX = 0.0 !
! D18 D(5,2,6,15) 67.0924 -DE/DX = 0.0 !
! D19 D(1,4,9,10) -116.2667 -DE/DX = 0.0 !
! D20 D(1,4,9,11) 124.6619 -DE/DX = 0.0 !
! D21 D(1,4,9,12) 9.1189 -DE/DX = 0.0 !
! D22 D(2,6,7,14) -107.3533 -DE/DX = 0.0 !
! D23 D(8,6,7,14) 104.5718 -DE/DX = 0.0 !
! D24 D(15,6,7,14) 4.057 -DE/DX = 0.0 !
! D25 D(2,6,15,14) 116.3168 -DE/DX = 0.0 !
! D26 D(7,6,15,14) -9.0638 -DE/DX = 0.0 !
! D27 D(8,6,15,14) -124.6141 -DE/DX = 0.0 !
! D28 D(6,7,14,10) 116.2997 -DE/DX = 0.0 !
! D29 D(6,7,14,15) -9.0629 -DE/DX = 0.0 !
! D30 D(6,7,14,16) -124.6244 -DE/DX = 0.0 !
! D31 D(4,9,10,13) -67.0954 -DE/DX = 0.0 !
! D32 D(4,9,10,14) 92.5996 -DE/DX = 0.0 !
! D33 D(11,9,10,13) 18.0958 -DE/DX = 0.0 !
! D34 D(11,9,10,14) 177.7907 -DE/DX = 0.0 !
! D35 D(12,9,10,13) 164.5001 -DE/DX = 0.0 !
! D36 D(12,9,10,14) -35.805 -DE/DX = 0.0 !
! D37 D(4,9,12,1) -4.0822 -DE/DX = 0.0 !
! D38 D(10,9,12,1) 107.3482 -DE/DX = 0.0 !
! D39 D(11,9,12,1) -104.5825 -DE/DX = 0.0 !
! D40 D(9,10,14,7) -92.6006 -DE/DX = 0.0 !
! D41 D(9,10,14,15) 35.7843 -DE/DX = 0.0 !
! D42 D(9,10,14,16) -177.758 -DE/DX = 0.0 !
! D43 D(13,10,14,7) 67.0945 -DE/DX = 0.0 !
! D44 D(13,10,14,15) -164.5206 -DE/DX = 0.0 !
! D45 D(13,10,14,16) -18.0629 -DE/DX = 0.0 !
! D46 D(7,14,15,6) 4.056 -DE/DX = 0.0 !
! D47 D(10,14,15,6) -107.3337 -DE/DX = 0.0 !
! D48 D(16,14,15,6) 104.553 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
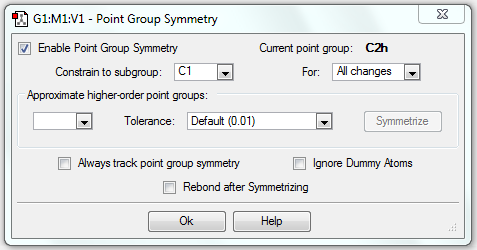
The point group of the optimised structure is C2h, confirming the structure is correct.
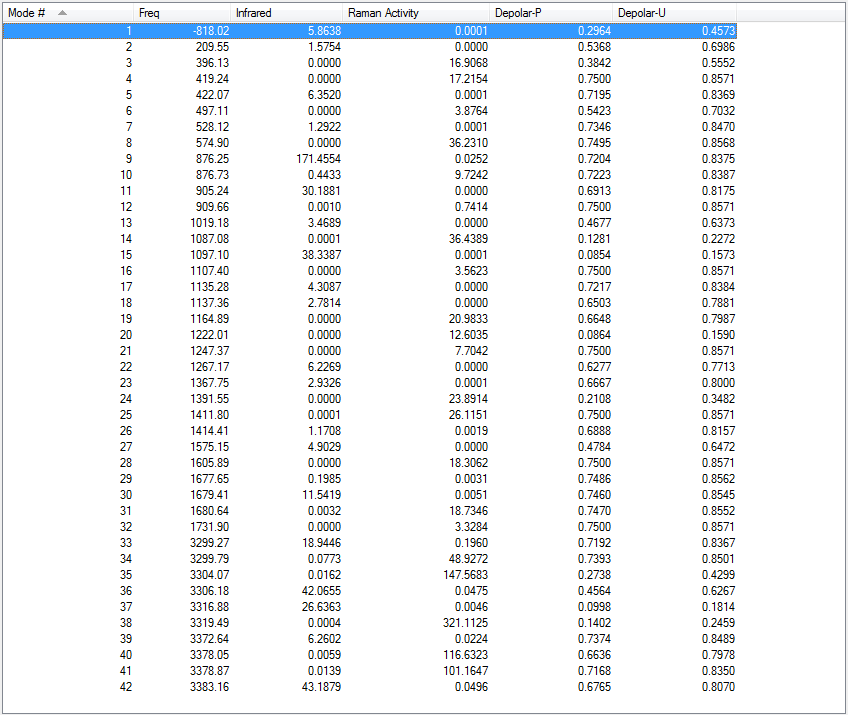
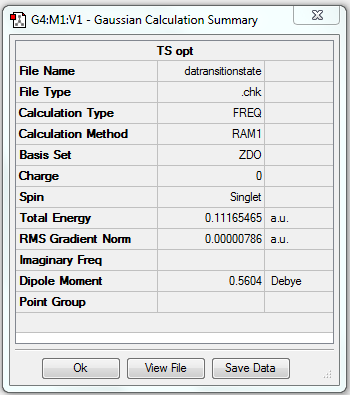
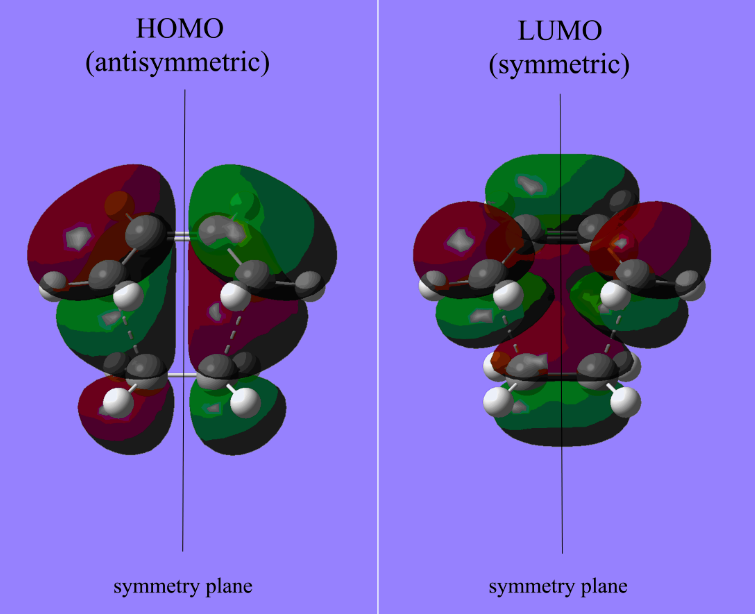
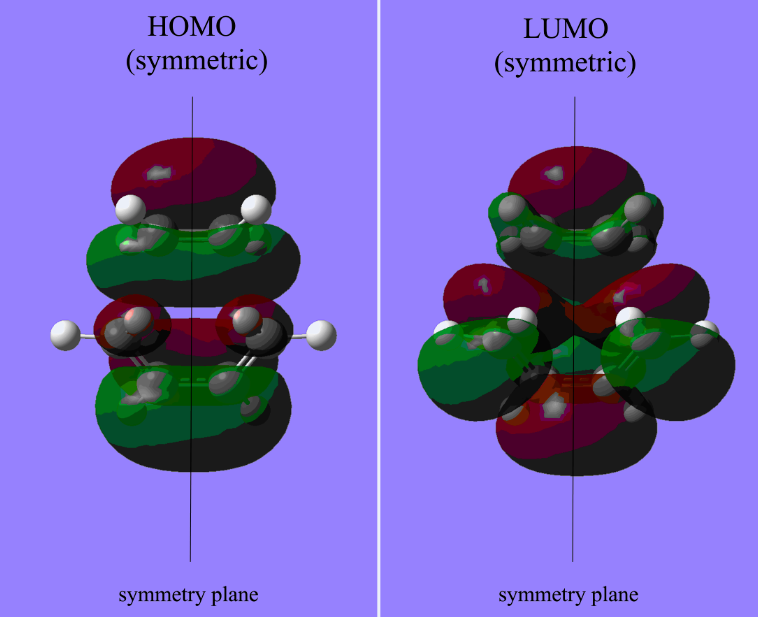
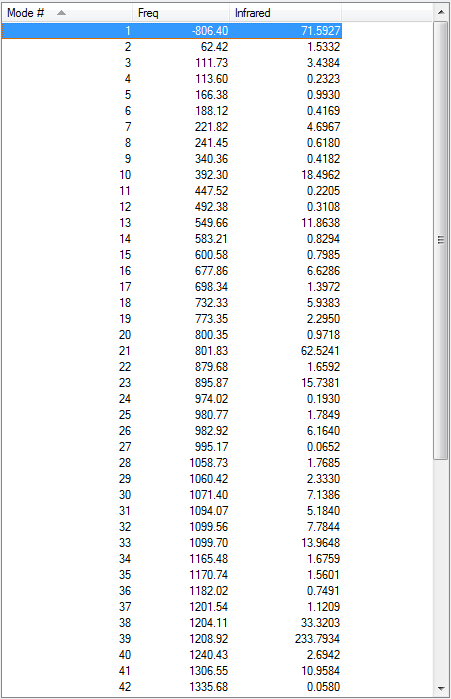
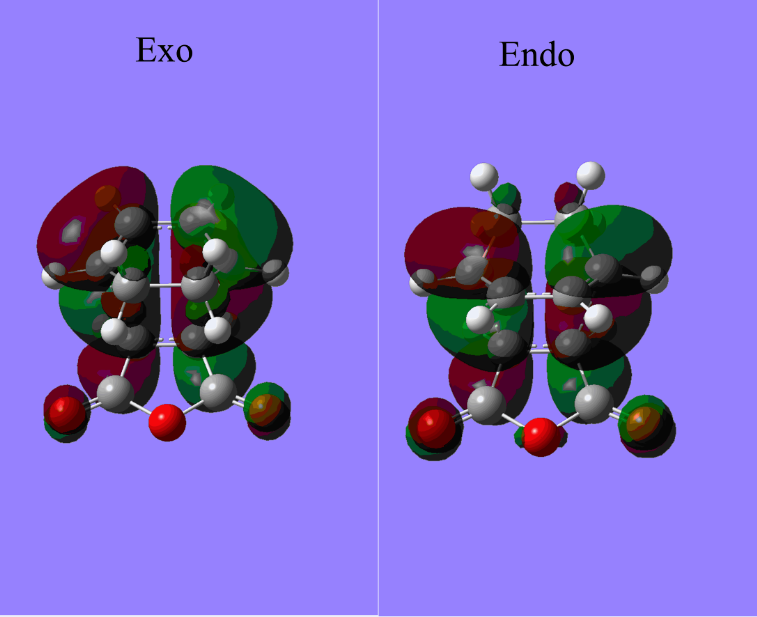
As the above table is shown, there is only one imaginary frequency that has a magnitude of 818.02 cm-1. The corresponding vibrational mode is shown below, in which two nearby terminal C atoms of the two allyl fragments approach each other in a sychronised motion and facilitates a bond formation, while the other pair of terminal C atoms move away from each other in a sychronised motion and represents a bond breaking. This is exactly what happens in the Cope rearrangement, that is, one C-C bond forms and one C-C bond breaks at the same time (i.e. a concerted process) in a 6-membered ring like system.
7. Thermochemistry
Sum of electronic and zero-point Energies= -231.466698 Sum of electronic and thermal Energies= -231.461339 Sum of electronic and thermal Enthalpies= -231.460395 Sum of electronic and thermal Free Energies= -231.495203
8. Key information
| Transition state type | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Chair | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | HF | 3-21G | Default | -231.61932237 a.u. | C2h |
(c) Optimisation of chair transition state using frozen coordinate method
(c)&(d) together is another way of optimisation which is more reliable and gives a better result, but more time-consuming.
1. Optimising chair transition state with frozen coordinates
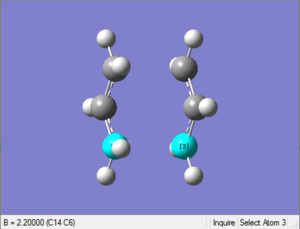
This optimised structure looks a lot like the one optimised in (b). However, as the GaussView image is shown on the right, the optimised structure has a fixed bond distance of 2.2 Å for the terminal C atoms of the allyl fragments.
2. File link
The optimised file is linked to here.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000030 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000514 0.001800 YES
RMS Displacement 0.000100 0.001200 YES
Predicted change in Energy=-2.609537D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.381 -DE/DX = 0.0005 !
! R2 R(1,3) 1.0742 -DE/DX = 0.0006 !
! R3 R(1,4) 1.0728 -DE/DX = 0.0003 !
! R4 R(1,9) 2.2112 -DE/DX = -0.0026 !
! R5 R(1,10) 2.794 -DE/DX = -0.0017 !
! R6 R(1,11) 2.6065 -DE/DX = -0.0014 !
! R7 R(1,12) 2.5199 -DE/DX = -0.0014 !
! R8 R(2,5) 1.076 -DE/DX = 0.0 !
! R9 R(2,6) 1.381 -DE/DX = 0.0005 !
! R10 R(2,9) 2.794 -DE/DX = -0.0017 !
! R11 R(2,14) 2.7939 -DE/DX = -0.0017 !
! R12 R(3,9) 2.6065 -DE/DX = -0.0014 !
! R13 R(4,9) 2.5199 -DE/DX = -0.0014 !
! R14 R(6,7) 1.0728 -DE/DX = 0.0003 !
! R15 R(6,8) 1.0742 -DE/DX = 0.0006 !
! R16 R(6,10) 2.7939 -DE/DX = -0.0017 !
! R17 R(6,14) 2.2111 -DE/DX = -0.0026 !
! R18 R(6,15) 2.5199 -DE/DX = -0.0014 !
! R19 R(6,16) 2.6064 -DE/DX = -0.0014 !
! R20 R(7,14) 2.5199 -DE/DX = -0.0014 !
! R21 R(8,14) 2.6064 -DE/DX = -0.0014 !
! R22 R(9,10) 1.3809 -DE/DX = 0.0005 !
! R23 R(9,11) 1.0742 -DE/DX = 0.0006 !
! R24 R(9,12) 1.0728 -DE/DX = 0.0003 !
! R25 R(10,13) 1.076 -DE/DX = 0.0 !
! R26 R(10,14) 1.381 -DE/DX = 0.0005 !
! R27 R(14,15) 1.0728 -DE/DX = 0.0003 !
! R28 R(14,16) 1.0742 -DE/DX = 0.0006 !
! A1 A(2,1,3) 120.0683 -DE/DX = -0.0002 !
! A2 A(2,1,4) 119.8179 -DE/DX = -0.0001 !
! A3 A(2,1,10) 82.4382 -DE/DX = 0.0 !
! A4 A(2,1,11) 123.5084 -DE/DX = 0.0004 !
! A5 A(2,1,12) 88.8549 -DE/DX = 0.0 !
! A6 A(3,1,4) 115.0816 -DE/DX = -0.0001 !
! A7 A(3,1,10) 128.2396 -DE/DX = 0.0006 !
! A8 A(3,1,11) 86.5797 -DE/DX = 0.0002 !
! A9 A(3,1,12) 84.9819 -DE/DX = 0.0002 !
! A10 A(4,1,10) 81.9499 -DE/DX = 0.0003 !
! A11 A(4,1,11) 80.5843 -DE/DX = 0.0002 !
! A12 A(4,1,12) 118.6613 -DE/DX = 0.0006 !
! A13 A(10,1,11) 46.3535 -DE/DX = 0.0004 !
! A14 A(10,1,12) 46.8798 -DE/DX = 0.0004 !
! A15 A(11,1,12) 41.3453 -DE/DX = 0.0004 !
! A16 A(1,2,5) 118.0882 -DE/DX = 0.0 !
! A17 A(1,2,6) 122.0681 -DE/DX = -0.0002 !
! A18 A(1,2,14) 97.5634 -DE/DX = 0.0 !
! A19 A(5,2,6) 118.0863 -DE/DX = 0.0 !
! A20 A(5,2,9) 108.7359 -DE/DX = 0.0001 !
! A21 A(5,2,14) 108.7302 -DE/DX = 0.0001 !
! A22 A(6,2,9) 97.5568 -DE/DX = 0.0 !
! A23 A(9,2,14) 51.2461 -DE/DX = 0.0005 !
! A24 A(2,6,7) 119.8147 -DE/DX = -0.0001 !
! A25 A(2,6,8) 120.0668 -DE/DX = -0.0002 !
! A26 A(2,6,10) 82.4415 -DE/DX = 0.0 !
! A27 A(2,6,15) 88.8495 -DE/DX = 0.0 !
! A28 A(2,6,16) 123.5089 -DE/DX = 0.0004 !
! A29 A(7,6,8) 115.0808 -DE/DX = -0.0001 !
! A30 A(7,6,10) 81.9518 -DE/DX = 0.0003 !
! A31 A(7,6,15) 118.6718 -DE/DX = 0.0006 !
! A32 A(7,6,16) 80.5978 -DE/DX = 0.0002 !
! A33 A(8,6,10) 128.2454 -DE/DX = 0.0006 !
! A34 A(8,6,15) 84.9896 -DE/DX = 0.0002 !
! A35 A(8,6,16) 86.5784 -DE/DX = 0.0002 !
! A36 A(10,6,15) 46.8812 -DE/DX = 0.0004 !
! A37 A(10,6,16) 46.356 -DE/DX = 0.0004 !
! A38 A(15,6,16) 41.3459 -DE/DX = 0.0004 !
! A39 A(2,9,3) 46.3531 -DE/DX = 0.0004 !
! A40 A(2,9,4) 46.8799 -DE/DX = 0.0004 !
! A41 A(2,9,10) 82.4378 -DE/DX = 0.0 !
! A42 A(2,9,11) 128.2377 -DE/DX = 0.0006 !
! A43 A(2,9,12) 81.9504 -DE/DX = 0.0003 !
! A44 A(3,9,4) 41.345 -DE/DX = 0.0004 !
! A45 A(3,9,10) 123.5075 -DE/DX = 0.0004 !
! A46 A(3,9,11) 86.5783 -DE/DX = 0.0002 !
! A47 A(3,9,12) 80.5853 -DE/DX = 0.0002 !
! A48 A(4,9,10) 88.8545 -DE/DX = 0.0 !
! A49 A(4,9,11) 84.98 -DE/DX = 0.0002 !
! A50 A(4,9,12) 118.6622 -DE/DX = 0.0006 !
! A51 A(10,9,11) 120.0689 -DE/DX = -0.0002 !
! A52 A(10,9,12) 119.8174 -DE/DX = -0.0001 !
! A53 A(11,9,12) 115.0822 -DE/DX = -0.0001 !
! A54 A(1,10,6) 51.2464 -DE/DX = 0.0005 !
! A55 A(1,10,13) 108.7341 -DE/DX = 0.0001 !
! A56 A(1,10,14) 97.5576 -DE/DX = 0.0 !
! A57 A(6,10,9) 97.5638 -DE/DX = 0.0 !
! A58 A(6,10,13) 108.7286 -DE/DX = 0.0001 !
! A59 A(9,10,13) 118.0879 -DE/DX = 0.0 !
! A60 A(9,10,14) 122.0682 -DE/DX = -0.0002 !
! A61 A(13,10,14) 118.0862 -DE/DX = 0.0 !
! A62 A(2,14,7) 46.8816 -DE/DX = 0.0004 !
! A63 A(2,14,8) 46.3559 -DE/DX = 0.0004 !
! A64 A(2,14,10) 82.4408 -DE/DX = 0.0 !
! A65 A(2,14,15) 81.9526 -DE/DX = 0.0003 !
! A66 A(2,14,16) 128.2454 -DE/DX = 0.0006 !
! A67 A(7,14,8) 41.3457 -DE/DX = 0.0004 !
! A68 A(7,14,10) 88.8491 -DE/DX = 0.0 !
! A69 A(7,14,15) 118.6737 -DE/DX = 0.0006 !
! A70 A(7,14,16) 84.9889 -DE/DX = 0.0002 !
! A71 A(8,14,10) 123.508 -DE/DX = 0.0004 !
! A72 A(8,14,15) 80.6001 -DE/DX = 0.0002 !
! A73 A(8,14,16) 86.5786 -DE/DX = 0.0002 !
! A74 A(10,14,15) 119.8129 -DE/DX = -0.0001 !
! A75 A(10,14,16) 120.0663 -DE/DX = -0.0002 !
! A76 A(15,14,16) 115.0826 -DE/DX = -0.0001 !
! D1 D(3,1,2,5) 14.1563 -DE/DX = 0.0005 !
! D2 D(3,1,2,6) 178.7583 -DE/DX = 0.0002 !
! D3 D(3,1,2,14) 130.135 -DE/DX = 0.0007 !
! D4 D(4,1,2,5) 167.8413 -DE/DX = -0.0005 !
! D5 D(4,1,2,6) -27.5568 -DE/DX = -0.0008 !
! D6 D(4,1,2,14) -76.1801 -DE/DX = -0.0003 !
! D7 D(10,1,2,5) -115.9806 -DE/DX = -0.0001 !
! D8 D(10,1,2,6) 48.6213 -DE/DX = -0.0004 !
! D9 D(10,1,2,14) -0.002 -DE/DX = 0.0 !
! D10 D(11,1,2,5) -93.3396 -DE/DX = 0.0001 !
! D11 D(11,1,2,6) 71.2624 -DE/DX = -0.0002 !
! D12 D(11,1,2,14) 22.6391 -DE/DX = 0.0002 !
! D13 D(12,1,2,5) -69.3747 -DE/DX = 0.0003 !
! D14 D(12,1,2,6) 95.2272 -DE/DX = 0.0 !
! D15 D(12,1,2,14) 46.6039 -DE/DX = 0.0004 !
! D16 D(2,1,10,6) -23.7685 -DE/DX = 0.0001 !
! D17 D(2,1,10,13) -123.1226 -DE/DX = -0.0001 !
! D18 D(2,1,10,14) 0.004 -DE/DX = 0.0 !
! D19 D(3,1,10,6) -146.3768 -DE/DX = 0.0001 !
! D20 D(3,1,10,13) 114.2691 -DE/DX = 0.0 !
! D21 D(3,1,10,14) -122.6043 -DE/DX = 0.0001 !
! D22 D(4,1,10,6) 97.9249 -DE/DX = -0.0001 !
! D23 D(4,1,10,13) -1.4292 -DE/DX = -0.0002 !
! D24 D(4,1,10,14) 121.6975 -DE/DX = -0.0001 !
! D25 D(11,1,10,6) -177.436 -DE/DX = 0.0 !
! D26 D(11,1,10,13) 83.2099 -DE/DX = -0.0001 !
! D27 D(11,1,10,14) -153.6635 -DE/DX = -0.0001 !
! D28 D(12,1,10,6) -119.3173 -DE/DX = 0.0001 !
! D29 D(12,1,10,13) 141.3286 -DE/DX = 0.0 !
! D30 D(12,1,10,14) -95.5447 -DE/DX = 0.0001 !
! D31 D(1,2,6,7) 27.5599 -DE/DX = 0.0008 !
! D32 D(1,2,6,8) -178.7689 -DE/DX = -0.0002 !
! D33 D(1,2,6,10) -48.6234 -DE/DX = 0.0004 !
! D34 D(1,2,6,15) -95.2319 -DE/DX = 0.0 !
! D35 D(1,2,6,16) -71.2759 -DE/DX = 0.0002 !
! D36 D(5,2,6,7) -167.8379 -DE/DX = 0.0005 !
! D37 D(5,2,6,8) -14.1667 -DE/DX = -0.0005 !
! D38 D(5,2,6,10) 115.9789 -DE/DX = 0.0001 !
! D39 D(5,2,6,15) 69.3703 -DE/DX = -0.0003 !
! D40 D(5,2,6,16) 93.3264 -DE/DX = -0.0001 !
! D41 D(9,2,6,7) 76.1817 -DE/DX = 0.0003 !
! D42 D(9,2,6,8) -130.1471 -DE/DX = -0.0007 !
! D43 D(9,2,6,10) -0.0016 -DE/DX = 0.0 !
! D44 D(9,2,6,15) -46.6102 -DE/DX = -0.0004 !
! D45 D(9,2,6,16) -22.6541 -DE/DX = -0.0002 !
! D46 D(5,2,9,3) 83.2092 -DE/DX = -0.0001 !
! D47 D(5,2,9,4) 141.3276 -DE/DX = 0.0 !
! D48 D(5,2,9,10) -123.1238 -DE/DX = -0.0001 !
! D49 D(5,2,9,11) 114.2682 -DE/DX = 0.0 !
! D50 D(5,2,9,12) -1.4309 -DE/DX = -0.0002 !
! D51 D(6,2,9,3) -153.6639 -DE/DX = -0.0001 !
! D52 D(6,2,9,4) -95.5455 -DE/DX = 0.0001 !
! D53 D(6,2,9,10) 0.0032 -DE/DX = 0.0 !
! D54 D(6,2,9,11) -122.6049 -DE/DX = 0.0001 !
! D55 D(6,2,9,12) 121.6961 -DE/DX = -0.0001 !
! D56 D(14,2,9,3) -177.436 -DE/DX = 0.0 !
! D57 D(14,2,9,4) -119.3176 -DE/DX = 0.0001 !
! D58 D(14,2,9,10) -23.769 -DE/DX = 0.0001 !
! D59 D(14,2,9,11) -146.3771 -DE/DX = 0.0001 !
! D60 D(14,2,9,12) 97.9239 -DE/DX = -0.0001 !
! D61 D(1,2,14,7) 95.5418 -DE/DX = -0.0001 !
! D62 D(1,2,14,8) 153.6589 -DE/DX = 0.0001 !
! D63 D(1,2,14,10) 0.004 -DE/DX = 0.0 !
! D64 D(1,2,14,15) -121.6829 -DE/DX = 0.0001 !
! D65 D(1,2,14,16) 122.614 -DE/DX = -0.0001 !
! D66 D(5,2,14,7) -141.328 -DE/DX = 0.0 !
! D67 D(5,2,14,8) -83.2109 -DE/DX = 0.0001 !
! D68 D(5,2,14,10) 123.1342 -DE/DX = 0.0001 !
! D69 D(5,2,14,15) 1.4472 -DE/DX = 0.0002 !
! D70 D(5,2,14,16) -114.2559 -DE/DX = 0.0 !
! D71 D(9,2,14,7) 119.3054 -DE/DX = -0.0001 !
! D72 D(9,2,14,8) 177.4225 -DE/DX = 0.0 !
! D73 D(9,2,14,10) 23.7676 -DE/DX = -0.0001 !
! D74 D(9,2,14,15) -97.9194 -DE/DX = 0.0001 !
! D75 D(9,2,14,16) 146.3775 -DE/DX = -0.0001 !
! D76 D(2,6,10,1) 23.7671 -DE/DX = -0.0001 !
! D77 D(2,6,10,9) 0.0032 -DE/DX = 0.0 !
! D78 D(2,6,10,13) 123.1327 -DE/DX = 0.0001 !
! D79 D(7,6,10,1) -97.9217 -DE/DX = 0.0001 !
! D80 D(7,6,10,9) -121.6857 -DE/DX = 0.0001 !
! D81 D(7,6,10,13) 1.4438 -DE/DX = 0.0002 !
! D82 D(8,6,10,1) 146.3782 -DE/DX = -0.0001 !
! D83 D(8,6,10,9) 122.6143 -DE/DX = -0.0001 !
! D84 D(8,6,10,13) -114.2562 -DE/DX = 0.0 !
! D85 D(15,6,10,1) 119.3049 -DE/DX = -0.0001 !
! D86 D(15,6,10,9) 95.541 -DE/DX = -0.0001 !
! D87 D(15,6,10,13) -141.3296 -DE/DX = 0.0 !
! D88 D(16,6,10,1) 177.4224 -DE/DX = 0.0 !
! D89 D(16,6,10,9) 153.6585 -DE/DX = 0.0001 !
! D90 D(16,6,10,13) -83.212 -DE/DX = 0.0001 !
! D91 D(2,9,10,6) -0.0016 -DE/DX = 0.0 !
! D92 D(2,9,10,13) -115.9785 -DE/DX = -0.0001 !
! D93 D(2,9,10,14) 48.6228 -DE/DX = -0.0004 !
! D94 D(3,9,10,6) 22.6395 -DE/DX = 0.0002 !
! D95 D(3,9,10,13) -93.3374 -DE/DX = 0.0001 !
! D96 D(3,9,10,14) 71.2639 -DE/DX = -0.0002 !
! D97 D(4,9,10,6) 46.6045 -DE/DX = 0.0004 !
! D98 D(4,9,10,13) -69.3724 -DE/DX = 0.0003 !
! D99 D(4,9,10,14) 95.2288 -DE/DX = 0.0 !
! D100 D(11,9,10,6) 130.133 -DE/DX = 0.0007 !
! D101 D(11,9,10,13) 14.1562 -DE/DX = 0.0005 !
! D102 D(11,9,10,14) 178.7574 -DE/DX = 0.0002 !
! D103 D(12,9,10,6) -76.1802 -DE/DX = -0.0003 !
! D104 D(12,9,10,13) 167.843 -DE/DX = -0.0005 !
! D105 D(12,9,10,14) -27.5558 -DE/DX = -0.0008 !
! D106 D(1,10,14,2) -0.002 -DE/DX = 0.0 !
! D107 D(1,10,14,7) -46.611 -DE/DX = -0.0004 !
! D108 D(1,10,14,8) -22.6546 -DE/DX = -0.0002 !
! D109 D(1,10,14,15) 76.1823 -DE/DX = 0.0003 !
! D110 D(1,10,14,16) -130.1469 -DE/DX = -0.0007 !
! D111 D(9,10,14,2) -48.6247 -DE/DX = 0.0004 !
! D112 D(9,10,14,7) -95.2337 -DE/DX = 0.0 !
! D113 D(9,10,14,8) -71.2774 -DE/DX = 0.0002 !
! D114 D(9,10,14,15) 27.5596 -DE/DX = 0.0008 !
! D115 D(9,10,14,16) -178.7696 -DE/DX = -0.0002 !
! D116 D(13,10,14,2) 115.9768 -DE/DX = 0.0001 !
! D117 D(13,10,14,7) 69.3678 -DE/DX = -0.0003 !
! D118 D(13,10,14,8) 93.3242 -DE/DX = -0.0001 !
! D119 D(13,10,14,15) -167.8389 -DE/DX = 0.0005 !
! D120 D(13,10,14,16) -14.1681 -DE/DX = -0.0005 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
The point group of the optimised structure is C2h, confirming the structure is correct.
6. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
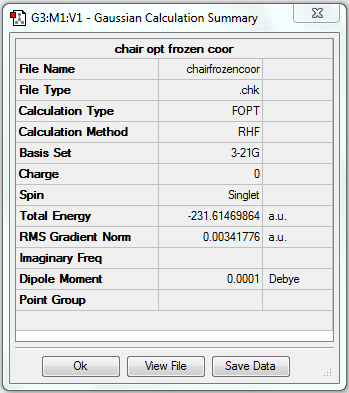
| Chair | Optimisation to a minimum | HF | 3-21G | Default | -231.61469864 a.u. | C2h |
(d) Reoptimisation of chair transition state with unfrozen coordinates
1. Optimising chair transition state using Berny method without force constants calculation
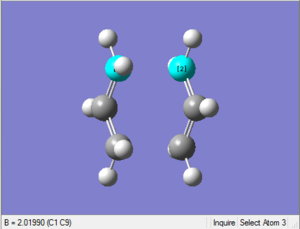
As the GaussView image is shown on the right, this optimised structure looks almost the same as the one optimised in (b).
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000027 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.000941 0.001800 YES
RMS Displacement 0.000181 0.001200 YES
Predicted change in Energy=-2.950432D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3893 -DE/DX = 0.0 !
! R2 R(1,3) 1.076 -DE/DX = 0.0 !
! R3 R(1,4) 1.0742 -DE/DX = 0.0 !
! R4 R(1,9) 2.0199 -DE/DX = 0.0 !
! R5 R(1,10) 2.6767 -DE/DX = 0.0 !
! R6 R(1,11) 2.457 -DE/DX = 0.0 !
! R7 R(1,12) 2.3914 -DE/DX = 0.0 !
! R8 R(2,5) 1.0759 -DE/DX = 0.0 !
! R9 R(2,6) 1.3893 -DE/DX = 0.0 !
! R10 R(2,9) 2.6767 -DE/DX = 0.0 !
! R11 R(2,12) 2.7767 -DE/DX = 0.0 !
! R12 R(2,14) 2.6767 -DE/DX = 0.0 !
! R13 R(2,15) 2.7766 -DE/DX = 0.0 !
! R14 R(3,9) 2.4569 -DE/DX = 0.0 !
! R15 R(4,9) 2.3914 -DE/DX = 0.0 !
! R16 R(4,10) 2.7767 -DE/DX = 0.0 !
! R17 R(6,7) 1.0742 -DE/DX = 0.0 !
! R18 R(6,8) 1.076 -DE/DX = 0.0 !
! R19 R(6,10) 2.6767 -DE/DX = 0.0 !
! R20 R(6,14) 2.0199 -DE/DX = 0.0 !
! R21 R(6,15) 2.3914 -DE/DX = 0.0 !
! R22 R(6,16) 2.4569 -DE/DX = 0.0 !
! R23 R(7,10) 2.7766 -DE/DX = 0.0 !
! R24 R(7,14) 2.3914 -DE/DX = 0.0 !
! R25 R(8,14) 2.457 -DE/DX = 0.0 !
! R26 R(9,10) 1.3893 -DE/DX = 0.0 !
! R27 R(9,11) 1.076 -DE/DX = 0.0 !
! R28 R(9,12) 1.0742 -DE/DX = 0.0 !
! R29 R(10,13) 1.0759 -DE/DX = 0.0 !
! R30 R(10,14) 1.3893 -DE/DX = 0.0 !
! R31 R(14,15) 1.0742 -DE/DX = 0.0 !
! R32 R(14,16) 1.076 -DE/DX = 0.0 !
! A1 A(2,1,3) 118.9978 -DE/DX = 0.0 !
! A2 A(2,1,4) 118.8761 -DE/DX = 0.0 !
! A3 A(2,1,10) 83.7831 -DE/DX = 0.0 !
! A4 A(2,1,11) 127.3466 -DE/DX = 0.0 !
! A5 A(3,1,4) 113.8178 -DE/DX = 0.0 !
! A6 A(3,1,10) 131.0861 -DE/DX = 0.0 !
! A7 A(3,1,11) 87.1004 -DE/DX = 0.0 !
! A8 A(3,1,12) 85.5397 -DE/DX = 0.0 !
! A9 A(4,1,11) 82.231 -DE/DX = 0.0 !
! A10 A(4,1,12) 122.6535 -DE/DX = 0.0 !
! A11 A(10,1,11) 48.7986 -DE/DX = 0.0 !
! A12 A(10,1,12) 49.2433 -DE/DX = 0.0 !
! A13 A(11,1,12) 43.5965 -DE/DX = 0.0 !
! A14 A(1,2,5) 118.1693 -DE/DX = 0.0 !
! A15 A(1,2,6) 120.5081 -DE/DX = 0.0 !
! A16 A(1,2,14) 96.2169 -DE/DX = 0.0 !
! A17 A(1,2,15) 106.9303 -DE/DX = 0.0 !
! A18 A(5,2,6) 118.1708 -DE/DX = 0.0 !
! A19 A(5,2,9) 109.3554 -DE/DX = 0.0 !
! A20 A(5,2,12) 86.755 -DE/DX = 0.0 !
! A21 A(5,2,14) 109.354 -DE/DX = 0.0 !
! A22 A(5,2,15) 86.7533 -DE/DX = 0.0 !
! A23 A(6,2,9) 96.2163 -DE/DX = 0.0 !
! A24 A(6,2,12) 106.9315 -DE/DX = 0.0 !
! A25 A(9,2,14) 53.5714 -DE/DX = 0.0 !
! A26 A(9,2,15) 59.4629 -DE/DX = 0.0 !
! A27 A(12,2,14) 59.4631 -DE/DX = 0.0 !
! A28 A(12,2,15) 54.8235 -DE/DX = 0.0 !
! A29 A(2,6,7) 118.877 -DE/DX = 0.0 !
! A30 A(2,6,8) 118.9972 -DE/DX = 0.0 !
! A31 A(2,6,10) 83.7837 -DE/DX = 0.0 !
! A32 A(2,6,16) 127.3461 -DE/DX = 0.0 !
! A33 A(7,6,8) 113.8153 -DE/DX = 0.0 !
! A34 A(7,6,15) 122.6532 -DE/DX = 0.0 !
! A35 A(7,6,16) 82.2318 -DE/DX = 0.0 !
! A36 A(8,6,10) 131.0929 -DE/DX = 0.0 !
! A37 A(8,6,15) 85.549 -DE/DX = 0.0 !
! A38 A(8,6,16) 87.1031 -DE/DX = 0.0 !
! A39 A(10,6,15) 49.2429 -DE/DX = 0.0 !
! A40 A(10,6,16) 48.7998 -DE/DX = 0.0 !
! A41 A(15,6,16) 43.5978 -DE/DX = 0.0 !
! A42 A(2,9,3) 48.7994 -DE/DX = 0.0 !
! A43 A(2,9,4) 49.2428 -DE/DX = 0.0 !
! A44 A(2,9,10) 83.7826 -DE/DX = 0.0 !
! A45 A(2,9,11) 131.0915 -DE/DX = 0.0 !
! A46 A(3,9,4) 43.5979 -DE/DX = 0.0 !
! A47 A(3,9,10) 127.3474 -DE/DX = 0.0 !
! A48 A(3,9,11) 87.1048 -DE/DX = 0.0 !
! A49 A(3,9,12) 82.2295 -DE/DX = 0.0 !
! A50 A(4,9,11) 85.5456 -DE/DX = 0.0 !
! A51 A(4,9,12) 122.6531 -DE/DX = 0.0 !
! A52 A(10,9,11) 118.9966 -DE/DX = 0.0 !
! A53 A(10,9,12) 118.8773 -DE/DX = 0.0 !
! A54 A(11,9,12) 113.8152 -DE/DX = 0.0 !
! A55 A(1,10,6) 53.5714 -DE/DX = 0.0 !
! A56 A(1,10,7) 59.4635 -DE/DX = 0.0 !
! A57 A(1,10,13) 109.3556 -DE/DX = 0.0 !
! A58 A(1,10,14) 96.2161 -DE/DX = 0.0 !
! A59 A(4,10,6) 59.4628 -DE/DX = 0.0 !
! A60 A(4,10,7) 54.8237 -DE/DX = 0.0 !
! A61 A(4,10,13) 86.7552 -DE/DX = 0.0 !
! A62 A(4,10,14) 106.9309 -DE/DX = 0.0 !
! A63 A(6,10,9) 96.2174 -DE/DX = 0.0 !
! A64 A(6,10,13) 109.3525 -DE/DX = 0.0 !
! A65 A(7,10,9) 106.9314 -DE/DX = 0.0 !
! A66 A(7,10,13) 86.7516 -DE/DX = 0.0 !
! A67 A(9,10,13) 118.1708 -DE/DX = 0.0 !
! A68 A(9,10,14) 120.5081 -DE/DX = 0.0 !
! A69 A(13,10,14) 118.1691 -DE/DX = 0.0 !
! A70 A(2,14,7) 49.2435 -DE/DX = 0.0 !
! A71 A(2,14,8) 48.7988 -DE/DX = 0.0 !
! A72 A(2,14,10) 83.784 -DE/DX = 0.0 !
! A73 A(2,14,16) 131.0869 -DE/DX = 0.0 !
! A74 A(7,14,8) 43.5964 -DE/DX = 0.0 !
! A75 A(7,14,15) 122.6545 -DE/DX = 0.0 !
! A76 A(7,14,16) 85.5425 -DE/DX = 0.0 !
! A77 A(8,14,10) 127.3449 -DE/DX = 0.0 !
! A78 A(8,14,15) 82.2341 -DE/DX = 0.0 !
! A79 A(8,14,16) 87.0983 -DE/DX = 0.0 !
! A80 A(10,14,15) 118.8758 -DE/DX = 0.0 !
! A81 A(10,14,16) 118.9984 -DE/DX = 0.0 !
! A82 A(15,14,16) 113.818 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 18.1822 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 177.7564 -DE/DX = 0.0 !
! D3 D(3,1,2,14) 134.0623 -DE/DX = 0.0 !
! D4 D(3,1,2,15) 113.6826 -DE/DX = 0.0 !
! D5 D(4,1,2,5) 164.5836 -DE/DX = 0.0 !
! D6 D(4,1,2,6) -35.8423 -DE/DX = 0.0 !
! D7 D(4,1,2,14) -79.5363 -DE/DX = 0.0 !
! D8 D(4,1,2,15) -99.9161 -DE/DX = 0.0 !
! D9 D(10,1,2,5) -115.8813 -DE/DX = 0.0 !
! D10 D(10,1,2,6) 43.6928 -DE/DX = 0.0 !
! D11 D(10,1,2,14) -0.0012 -DE/DX = 0.0 !
! D12 D(10,1,2,15) -20.381 -DE/DX = 0.0 !
! D13 D(11,1,2,5) -92.3169 -DE/DX = 0.0 !
! D14 D(11,1,2,6) 67.2573 -DE/DX = 0.0 !
! D15 D(11,1,2,14) 23.5632 -DE/DX = 0.0 !
! D16 D(11,1,2,15) 3.1835 -DE/DX = 0.0 !
! D17 D(2,1,10,6) -22.5778 -DE/DX = 0.0 !
! D18 D(2,1,10,7) -48.8671 -DE/DX = 0.0 !
! D19 D(2,1,10,13) -122.7886 -DE/DX = 0.0 !
! D20 D(2,1,10,14) 0.0024 -DE/DX = 0.0 !
! D21 D(3,1,10,6) -146.082 -DE/DX = 0.0 !
! D22 D(3,1,10,7) -172.3714 -DE/DX = 0.0 !
! D23 D(3,1,10,13) 113.7072 -DE/DX = 0.0 !
! D24 D(3,1,10,14) -123.5018 -DE/DX = 0.0 !
! D25 D(11,1,10,6) -177.5912 -DE/DX = 0.0 !
! D26 D(11,1,10,7) 156.1194 -DE/DX = 0.0 !
! D27 D(11,1,10,13) 82.198 -DE/DX = 0.0 !
! D28 D(11,1,10,14) -155.0111 -DE/DX = 0.0 !
! D29 D(12,1,10,6) -118.6663 -DE/DX = 0.0 !
! D30 D(12,1,10,7) -144.9557 -DE/DX = 0.0 !
! D31 D(12,1,10,13) 141.1229 -DE/DX = 0.0 !
! D32 D(12,1,10,14) -96.0861 -DE/DX = 0.0 !
! D33 D(1,2,6,7) 35.8386 -DE/DX = 0.0 !
! D34 D(1,2,6,8) -177.765 -DE/DX = 0.0 !
! D35 D(1,2,6,10) -43.693 -DE/DX = 0.0 !
! D36 D(1,2,6,16) -67.2627 -DE/DX = 0.0 !
! D37 D(5,2,6,7) -164.5875 -DE/DX = 0.0 !
! D38 D(5,2,6,8) -18.1911 -DE/DX = 0.0 !
! D39 D(5,2,6,10) 115.8808 -DE/DX = 0.0 !
! D40 D(5,2,6,16) 92.3112 -DE/DX = 0.0 !
! D41 D(9,2,6,7) 79.5305 -DE/DX = 0.0 !
! D42 D(9,2,6,8) -134.073 -DE/DX = 0.0 !
! D43 D(9,2,6,10) -0.0011 -DE/DX = 0.0 !
! D44 D(9,2,6,16) -23.5707 -DE/DX = 0.0 !
! D45 D(12,2,6,7) 99.9088 -DE/DX = 0.0 !
! D46 D(12,2,6,8) -113.6948 -DE/DX = 0.0 !
! D47 D(12,2,6,10) 20.3771 -DE/DX = 0.0 !
! D48 D(12,2,6,16) -3.1925 -DE/DX = 0.0 !
! D49 D(5,2,9,3) 82.1946 -DE/DX = 0.0 !
! D50 D(5,2,9,4) 141.1213 -DE/DX = 0.0 !
! D51 D(5,2,9,10) -122.7908 -DE/DX = 0.0 !
! D52 D(5,2,9,11) 113.7046 -DE/DX = 0.0 !
! D53 D(6,2,9,3) -155.0125 -DE/DX = 0.0 !
! D54 D(6,2,9,4) -96.0857 -DE/DX = 0.0 !
! D55 D(6,2,9,10) 0.0021 -DE/DX = 0.0 !
! D56 D(6,2,9,11) -123.5024 -DE/DX = 0.0 !
! D57 D(14,2,9,3) -177.5926 -DE/DX = 0.0 !
! D58 D(14,2,9,4) -118.6659 -DE/DX = 0.0 !
! D59 D(14,2,9,10) -22.578 -DE/DX = 0.0 !
! D60 D(14,2,9,11) -146.0825 -DE/DX = 0.0 !
! D61 D(15,2,9,3) 156.1179 -DE/DX = 0.0 !
! D62 D(15,2,9,4) -144.9554 -DE/DX = 0.0 !
! D63 D(15,2,9,10) -48.8675 -DE/DX = 0.0 !
! D64 D(15,2,9,11) -172.372 -DE/DX = 0.0 !
! D65 D(9,2,12,1) -51.7445 -DE/DX = 0.0 !
! D66 D(1,2,14,7) 96.0846 -DE/DX = 0.0 !
! D67 D(1,2,14,8) 155.0091 -DE/DX = 0.0 !
! D68 D(1,2,14,10) 0.0024 -DE/DX = 0.0 !
! D69 D(1,2,14,16) 123.5086 -DE/DX = 0.0 !
! D70 D(5,2,14,7) -141.1243 -DE/DX = 0.0 !
! D71 D(5,2,14,8) -82.1998 -DE/DX = 0.0 !
! D72 D(5,2,14,10) 122.7935 -DE/DX = 0.0 !
! D73 D(5,2,14,16) -113.7003 -DE/DX = 0.0 !
! D74 D(9,2,14,7) 118.6602 -DE/DX = 0.0 !
! D75 D(9,2,14,8) 177.5847 -DE/DX = 0.0 !
! D76 D(9,2,14,10) 22.5779 -DE/DX = 0.0 !
! D77 D(9,2,14,16) 146.0842 -DE/DX = 0.0 !
! D78 D(12,2,14,7) 144.949 -DE/DX = 0.0 !
! D79 D(12,2,14,8) -156.1265 -DE/DX = 0.0 !
! D80 D(12,2,14,10) 48.8667 -DE/DX = 0.0 !
! D81 D(12,2,14,16) 172.373 -DE/DX = 0.0 !
! D82 D(1,4,9,10) -116.2603 -DE/DX = 0.0 !
! D83 D(2,6,10,1) 22.5779 -DE/DX = 0.0 !
! D84 D(2,6,10,4) 48.8668 -DE/DX = 0.0 !
! D85 D(2,6,10,9) 0.0021 -DE/DX = 0.0 !
! D86 D(2,6,10,13) 122.7946 -DE/DX = 0.0 !
! D87 D(8,6,10,1) 146.0849 -DE/DX = 0.0 !
! D88 D(8,6,10,4) 172.3738 -DE/DX = 0.0 !
! D89 D(8,6,10,9) 123.5092 -DE/DX = 0.0 !
! D90 D(8,6,10,13) -113.6983 -DE/DX = 0.0 !
! D91 D(15,6,10,1) 118.6598 -DE/DX = 0.0 !
! D92 D(15,6,10,4) 144.9487 -DE/DX = 0.0 !
! D93 D(15,6,10,9) 96.084 -DE/DX = 0.0 !
! D94 D(15,6,10,13) -141.1234 -DE/DX = 0.0 !
! D95 D(16,6,10,1) 177.5861 -DE/DX = 0.0 !
! D96 D(16,6,10,4) -156.125 -DE/DX = 0.0 !
! D97 D(16,6,10,9) 155.0103 -DE/DX = 0.0 !
! D98 D(16,6,10,13) -82.1971 -DE/DX = 0.0 !
! D99 D(14,6,15,2) -116.2658 -DE/DX = 0.0 !
! D100 D(6,7,10,14) -51.7402 -DE/DX = 0.0 !
! D101 D(2,9,10,6) -0.0011 -DE/DX = 0.0 !
! D102 D(2,9,10,7) -20.3807 -DE/DX = 0.0 !
! D103 D(2,9,10,13) -115.8803 -DE/DX = 0.0 !
! D104 D(2,9,10,14) 43.6931 -DE/DX = 0.0 !
! D105 D(3,9,10,6) 23.5628 -DE/DX = 0.0 !
! D106 D(3,9,10,7) 3.1832 -DE/DX = 0.0 !
! D107 D(3,9,10,13) -92.3164 -DE/DX = 0.0 !
! D108 D(3,9,10,14) 67.257 -DE/DX = 0.0 !
! D109 D(11,9,10,6) 134.0682 -DE/DX = 0.0 !
! D110 D(11,9,10,7) 113.6885 -DE/DX = 0.0 !
! D111 D(11,9,10,13) 18.189 -DE/DX = 0.0 !
! D112 D(11,9,10,14) 177.7623 -DE/DX = 0.0 !
! D113 D(12,9,10,6) -79.5361 -DE/DX = 0.0 !
! D114 D(12,9,10,7) -99.9157 -DE/DX = 0.0 !
! D115 D(12,9,10,13) 164.5847 -DE/DX = 0.0 !
! D116 D(12,9,10,14) -35.8419 -DE/DX = 0.0 !
! D117 D(1,10,14,2) -0.0012 -DE/DX = 0.0 !
! D118 D(1,10,14,8) -23.5715 -DE/DX = 0.0 !
! D119 D(1,10,14,15) 79.5312 -DE/DX = 0.0 !
! D120 D(1,10,14,16) -134.0665 -DE/DX = 0.0 !
! D121 D(4,10,14,2) 20.3771 -DE/DX = 0.0 !
! D122 D(4,10,14,8) -3.1931 -DE/DX = 0.0 !
! D123 D(4,10,14,15) 99.9095 -DE/DX = 0.0 !
! D124 D(4,10,14,16) -113.6881 -DE/DX = 0.0 !
! D125 D(9,10,14,2) -43.6936 -DE/DX = 0.0 !
! D126 D(9,10,14,8) -67.2639 -DE/DX = 0.0 !
! D127 D(9,10,14,15) 35.8388 -DE/DX = 0.0 !
! D128 D(9,10,14,16) -177.7589 -DE/DX = 0.0 !
! D129 D(13,10,14,2) 115.8801 -DE/DX = 0.0 !
! D130 D(13,10,14,8) 92.3099 -DE/DX = 0.0 !
! D131 D(13,10,14,15) -164.5875 -DE/DX = 0.0 !
! D132 D(13,10,14,16) -18.1852 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
The point group of the optimised structure is C2h, confirming the structure is correct.
Smilarly to (b), there is also only one imaginary frequency of about -818 cm-1. The corresponding vibrational mode is shown above, and the motion is similar to that of the structure obtained in (b).
7. Thermochemistry
Sum of electronic and zero-point Energies= -231.466696 Sum of electronic and thermal Energies= -231.461337 Sum of electronic and thermal Enthalpies= -231.460392 Sum of electronic and thermal Free Energies= -231.495201
8. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
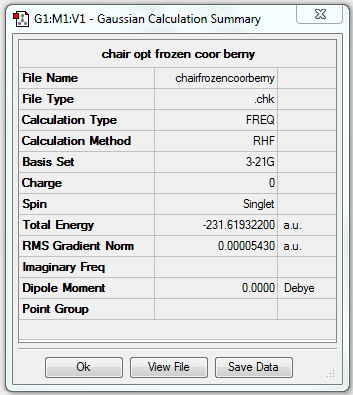
| Chair | Optimisation to a TS (Berny) | HF | 3-21G | Default | -231.61932200 a.u. | C2h |
9. Comparison to (b)
| Bond forming/breaking distances (b) | Bond forming/breaking distances (d) |
|---|---|
| 2.02038 Å | 2.01990 Å |
The bond forming/breaking distances of the optimised structure using the redundant coordinate editor are just slightly lower than that of the optimised structure by computing the force constants.
(e) Optimisation of boat transition state using QST2 method
First optimisation from optimised anti2 1,5-hexadiene
1. Optimising boat transition state from optimised anti2 1,5-hexadiene
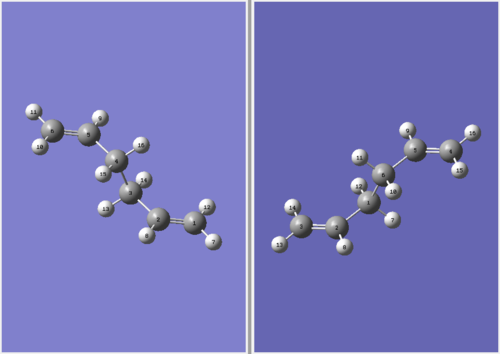
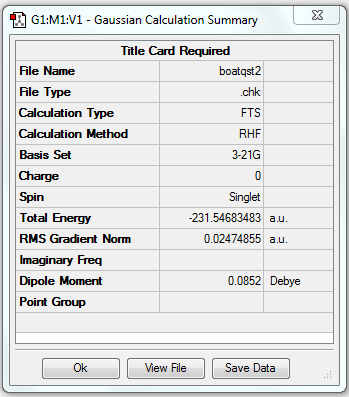
The QST2 method does not give the correct boat structure using these reactant and product structures shown above. The optimisation failed and we obtained the structure shown below which looks a bit like the chair transition state but more dissociated.
test molecule |
2. File link
The optimised file is linked to here.
The gradient is larger than 0.001, which means the optimisation is not complete.
The point group of the optimised structure is C2, indicating the structure is incorrect.
5. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Chair-like | Optimisation to a TS (QST2) | HF | 3-21G | Default | -231.54683483 a.u. | C2 |
Second optimisation from modified reactant and product
1. Optimising boat transition state from modified reactant and product

As the above GaussView image is shown, the reactant and product structures are modified to become closer to the boat transition state so that it is easier to obtain the correct boat structure.
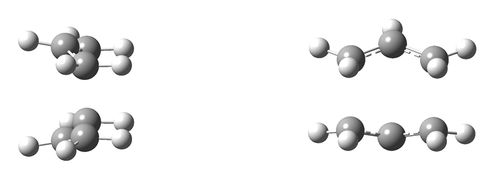
test molecule |
The optimised structure shown above is boat-like and is very similar to the one in Appendix 2 shown on the right.
2. File link
The optimised file is linked to here.
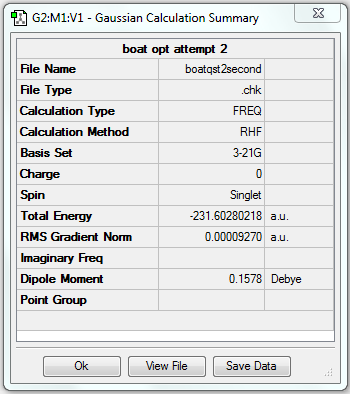
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000133 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.001256 0.001800 YES
RMS Displacement 0.000270 0.001200 YES
Predicted change in Energy=-2.879547D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition TS Reactant Product Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3815 1.3161 1.509 -DE/DX = 0.0 !
! R2 R(1,6) 2.1404 3.2253 1.5526 -DE/DX = -0.0001 !
! R3 R(1,7) 1.0739 1.0734 1.0856 -DE/DX = 0.0 !
! R4 R(1,12) 1.0742 1.0746 1.0848 -DE/DX = 0.0 !
! R5 R(2,3) 1.3813 1.509 1.3161 -DE/DX = -0.0001 !
! R6 R(2,8) 1.0764 1.0769 1.0769 -DE/DX = 0.0 !
! R7 R(3,4) 2.1418 1.5526 3.2253 -DE/DX = 0.0001 !
! R8 R(3,13) 1.0739 1.0856 1.0734 -DE/DX = 0.0 !
! R9 R(3,14) 1.0742 1.0848 1.0746 -DE/DX = 0.0 !
! R10 R(4,5) 1.3813 1.509 1.3161 -DE/DX = -0.0001 !
! R11 R(4,15) 1.0742 1.0848 1.0746 -DE/DX = 0.0 !
! R12 R(4,16) 1.0739 1.0856 1.0734 -DE/DX = 0.0 !
! R13 R(5,6) 1.3815 1.3161 1.509 -DE/DX = 0.0 !
! R14 R(5,9) 1.0764 1.0769 1.0769 -DE/DX = 0.0 !
! R15 R(6,10) 1.0742 1.0746 1.0848 -DE/DX = 0.0 !
! R16 R(6,11) 1.0739 1.0734 1.0856 -DE/DX = 0.0 !
! A1 A(2,1,6) 103.3627 64.1277 100.0 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.6476 121.8643 112.7378 -DE/DX = 0.0 !
! A3 A(2,1,12) 118.859 121.822 112.8447 -DE/DX = 0.0 !
! A4 A(6,1,7) 101.0441 98.0896 111.2032 -DE/DX = 0.0 !
! A5 A(6,1,12) 91.3879 108.832 112.3204 -DE/DX = 0.0 !
! A6 A(7,1,12) 114.7026 116.3134 107.7104 -DE/DX = 0.0 !
! A7 A(1,2,3) 121.6901 124.8104 124.8104 -DE/DX = 0.0 !
! A8 A(1,2,8) 117.464 119.6718 115.5094 -DE/DX = 0.0 !
! A9 A(3,2,8) 117.4633 115.5094 119.6718 -DE/DX = 0.0 !
! A10 A(2,3,4) 103.336 100.0 64.1277 -DE/DX = 0.0 !
! A11 A(2,3,13) 119.6576 112.7378 121.8643 -DE/DX = 0.0 !
! A12 A(2,3,14) 118.8859 112.8447 121.822 -DE/DX = 0.0 !
! A13 A(4,3,13) 101.0147 111.2032 98.0896 -DE/DX = 0.0 !
! A14 A(4,3,14) 91.3529 112.3204 108.832 -DE/DX = 0.0 !
! A15 A(13,3,14) 114.7123 107.7104 116.3134 -DE/DX = 0.0 !
! A16 A(3,4,5) 103.336 100.0 64.1277 -DE/DX = 0.0 !
! A17 A(3,4,15) 91.3529 112.3204 108.832 -DE/DX = 0.0 !
! A18 A(3,4,16) 101.0147 111.2032 98.0896 -DE/DX = 0.0 !
! A19 A(5,4,15) 118.8859 112.8447 121.822 -DE/DX = 0.0 !
! A20 A(5,4,16) 119.6576 112.7378 121.8643 -DE/DX = 0.0 !
! A21 A(15,4,16) 114.7123 107.7104 116.3134 -DE/DX = 0.0 !
! A22 A(4,5,6) 121.6901 124.8104 124.8104 -DE/DX = 0.0 !
! A23 A(4,5,9) 117.4633 115.5094 119.6718 -DE/DX = 0.0 !
! A24 A(6,5,9) 117.464 119.6718 115.5094 -DE/DX = 0.0 !
! A25 A(1,6,5) 103.3627 64.1277 100.0 -DE/DX = 0.0 !
! A26 A(1,6,10) 91.3879 108.832 112.3204 -DE/DX = 0.0 !
! A27 A(1,6,11) 101.0441 98.0896 111.2032 -DE/DX = 0.0 !
! A28 A(5,6,10) 118.859 121.822 112.8447 -DE/DX = 0.0 !
! A29 A(5,6,11) 119.6476 121.8643 112.7378 -DE/DX = 0.0 !
! A30 A(10,6,11) 114.7026 116.3134 107.7104 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 64.8318 95.8625 114.6503 -DE/DX = 0.0 !
! D2 D(6,1,2,8) -93.873 -83.0289 -64.2825 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 176.0402 179.126 -127.1791 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 17.3354 0.2346 53.8882 -DE/DX = 0.0001 !
! D5 D(12,1,2,3) -34.3366 -1.1023 -4.8714 -DE/DX = -0.0001 !
! D6 D(12,1,2,8) 166.9586 -179.9937 176.1959 -DE/DX = 0.0 !
! D7 D(2,1,6,5) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D8 D(2,1,6,10) -120.1316 -116.9853 -119.9037 -DE/DX = 0.0 !
! D9 D(2,1,6,11) 124.3597 121.5795 119.2998 -DE/DX = 0.0 !
! D10 D(7,1,6,5) -124.3597 -121.5795 -119.2998 -DE/DX = 0.0 !
! D11 D(7,1,6,10) 115.5087 121.4353 120.7965 -DE/DX = 0.0 !
! D12 D(7,1,6,11) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D13 D(12,1,6,5) 120.1316 116.9853 119.9037 -DE/DX = 0.0 !
! D14 D(12,1,6,10) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D15 D(12,1,6,11) -115.5087 -121.4353 -120.7965 -DE/DX = 0.0 !
! D16 D(1,2,3,4) -64.8183 -114.6503 -95.8625 -DE/DX = 0.0 !
! D17 D(1,2,3,13) -175.9761 127.1791 -179.126 -DE/DX = 0.0 !
! D18 D(1,2,3,14) 34.3014 4.8714 1.1023 -DE/DX = 0.0 !
! D19 D(8,2,3,4) 93.8867 64.2825 83.0289 -DE/DX = 0.0 !
! D20 D(8,2,3,13) -17.2712 -53.8882 -0.2346 -DE/DX = 0.0 !
! D21 D(8,2,3,14) -166.9936 -176.1959 179.9937 -DE/DX = 0.0 !
! D22 D(2,3,4,5) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D23 D(2,3,4,15) 120.1451 119.9037 116.9853 -DE/DX = 0.0 !
! D24 D(2,3,4,16) -124.3478 -119.2998 -121.5795 -DE/DX = 0.0 !
! D25 D(13,3,4,5) 124.3478 119.2998 121.5795 -DE/DX = 0.0 !
! D26 D(13,3,4,15) -115.5071 -120.7965 -121.4353 -DE/DX = 0.0 !
! D27 D(13,3,4,16) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D28 D(14,3,4,5) -120.1451 -119.9037 -116.9853 -DE/DX = 0.0 !
! D29 D(14,3,4,15) 0.0 0.0 0.0 -DE/DX = 0.0 !
! D30 D(14,3,4,16) 115.5071 120.7965 121.4353 -DE/DX = 0.0 !
! D31 D(3,4,5,6) 64.8183 114.6503 95.8625 -DE/DX = 0.0 !
! D32 D(3,4,5,9) -93.8867 -64.2825 -83.0289 -DE/DX = 0.0 !
! D33 D(15,4,5,6) -34.3014 -4.8714 -1.1023 -DE/DX = 0.0 !
! D34 D(15,4,5,9) 166.9936 176.1959 -179.9937 -DE/DX = 0.0 !
! D35 D(16,4,5,6) 175.9761 -127.1791 179.126 -DE/DX = 0.0 !
! D36 D(16,4,5,9) 17.2712 53.8882 0.2346 -DE/DX = 0.0 !
! D37 D(4,5,6,1) -64.8318 -95.8625 -114.6503 -DE/DX = 0.0 !
! D38 D(4,5,6,10) 34.3366 1.1023 4.8714 -DE/DX = 0.0001 !
! D39 D(4,5,6,11) -176.0402 -179.126 127.1791 -DE/DX = 0.0 !
! D40 D(9,5,6,1) 93.873 83.0289 64.2825 -DE/DX = 0.0 !
! D41 D(9,5,6,10) -166.9586 179.9937 -176.1959 -DE/DX = 0.0 !
! D42 D(9,5,6,11) -17.3354 -0.2346 -53.8882 -DE/DX = -0.0001 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
The point group of the optimised structure is C2v, confirming the structure is correct.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 839.62 cm-1. The corresponding vibrational mode is shown below. Although the geometry is different, the boat structure has a similar vibrational motion as the chair structure obtained earlier, that is, two nearby terminal C atoms of the two allyl fragments approach each other in a sychronised motion and facilitates a bond formation, while the other pair of terminal C atoms move away from each other in a sychronised motion and represents a bond breaking. Again, the bond forming and breaking happens at the same time.
7. Thermochemistry
Sum of electronic and zero-point Energies= -231.450934 Sum of electronic and thermal Energies= -231.445305 Sum of electronic and thermal Enthalpies= -231.444361 Sum of electronic and thermal Free Energies= -231.479780
8. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Boat | Optimisation to a TS (QST2) | HF | 3-21G | Default | -231.60280218 a.u. | C2v |
(f) IRC analysis of optimised chair and boat transition states
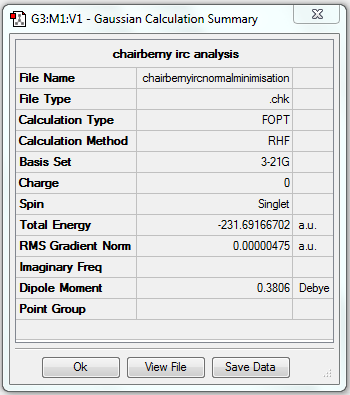
IRC analysis of optimised chair transition state
1. Calculating minimum energy path from chair transition state
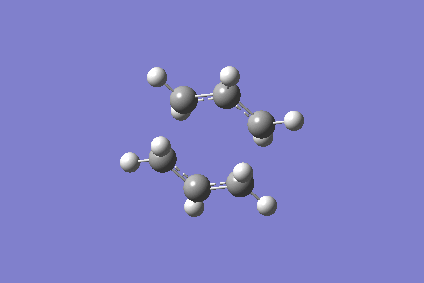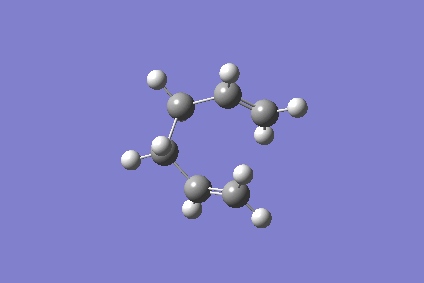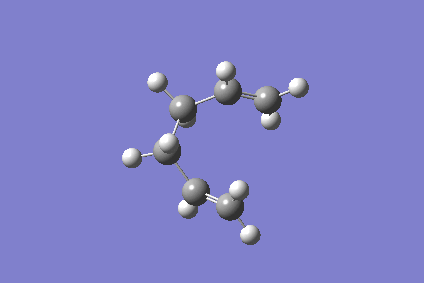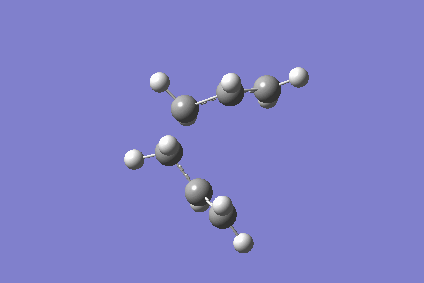
As the reaction coordinate is symmetrical in the cope rearrangement, "forward only" is chosen for this IRC calculation. There are 44 intermediate geometries obtianed, which are connected together to show the geometric change following the calculated minimum energy path from the boat transition structure to either reactant or product.
2. File link
The IRC analysed file is linked to here.
3. Structure of the last point of the IRC calculation
test molecule |
4. General information of the last point of the IRC calculation
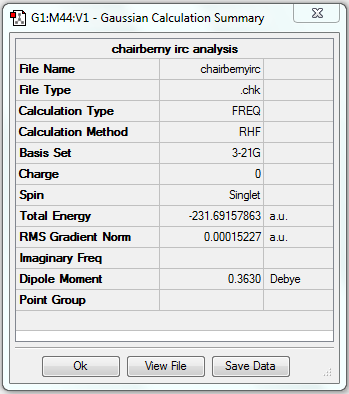
The gradient is less than 0.001, which means the optimisation is complete.
5. Symmetry information of the last point of the IRC calculation
6. Key information of the IRC calculation
| Transition state type | Job type | Method | Basis set | Memory limit | Energy of the last point | Point group of the last point |
|---|---|---|---|---|---|---|
| Chair | IRC, forward only, calculate always, compute 50 points | HF | 3-21G | Default | -231.69157863 a.u. | C2 |
7. IRC plot of the IRC calculation
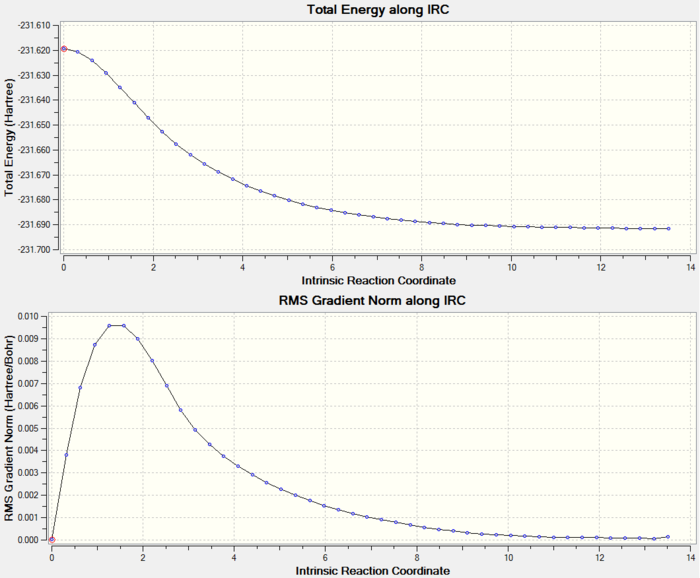
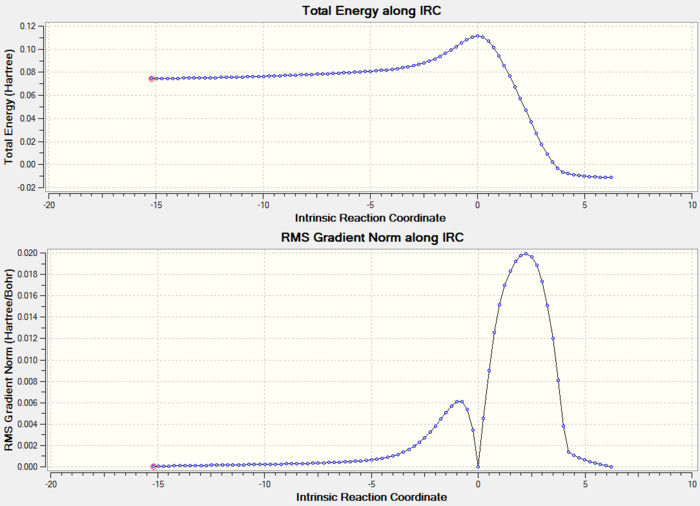
As the IRC plot is shown above, the energy minimum is not reached in this calculation because the RMS gradient does not reach 0 in the end. Now there are three options can be used to give a more accurate and reliable result: (1) run normal minimisation on the last point of this IRC calculation, (2) redo the IRC calculation to compute a larger number of points until the minimum is reached, (3) redo the IRC calculation to compute the force constants at every step. As the RMS gradient is very close to 0 already, I decided to try the first approach in the following.
(1) Normal minimisation
1. Optimising the last point of the IRC calculation using HF/3-21G
test molecule |
The structure looks almost the same as the one before optimisation.
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete. And the energy is the minimum I found, which is only slightly lower than that before optimisation.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000010 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000297 0.001800 YES
RMS Displacement 0.000090 0.001200 YES
Predicted change in Energy=-2.412823D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5083 -DE/DX = 0.0 !
! R2 R(1,3) 1.087 -DE/DX = 0.0 !
! R3 R(1,4) 1.0849 -DE/DX = 0.0 !
! R4 R(1,9) 1.5508 -DE/DX = 0.0 !
! R5 R(2,5) 1.0768 -DE/DX = 0.0 !
! R6 R(2,6) 1.3157 -DE/DX = 0.0 !
! R7 R(6,7) 1.0746 -DE/DX = 0.0 !
! R8 R(6,8) 1.0733 -DE/DX = 0.0 !
! R9 R(9,10) 1.5083 -DE/DX = 0.0 !
! R10 R(9,11) 1.087 -DE/DX = 0.0 !
! R11 R(9,12) 1.0849 -DE/DX = 0.0 !
! R12 R(10,13) 1.0768 -DE/DX = 0.0 !
! R13 R(10,14) 1.3157 -DE/DX = 0.0 !
! R14 R(14,15) 1.0746 -DE/DX = 0.0 !
! R15 R(14,16) 1.0733 -DE/DX = 0.0 !
! A1 A(2,1,3) 109.2943 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.9836 -DE/DX = 0.0 !
! A3 A(2,1,9) 112.0405 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.4615 -DE/DX = 0.0 !
! A5 A(3,1,9) 108.389 -DE/DX = 0.0 !
! A6 A(4,1,9) 109.5498 -DE/DX = 0.0 !
! A7 A(1,2,5) 115.3201 -DE/DX = 0.0 !
! A8 A(1,2,6) 124.9751 -DE/DX = 0.0 !
! A9 A(5,2,6) 119.7048 -DE/DX = 0.0 !
! A10 A(2,6,7) 121.8623 -DE/DX = 0.0 !
! A11 A(2,6,8) 121.8382 -DE/DX = 0.0 !
! A12 A(7,6,8) 116.2993 -DE/DX = 0.0 !
! A13 A(1,9,10) 112.0405 -DE/DX = 0.0 !
! A14 A(1,9,11) 108.389 -DE/DX = 0.0 !
! A15 A(1,9,12) 109.5498 -DE/DX = 0.0 !
! A16 A(10,9,11) 109.2943 -DE/DX = 0.0 !
! A17 A(10,9,12) 109.9836 -DE/DX = 0.0 !
! A18 A(11,9,12) 107.4615 -DE/DX = 0.0 !
! A19 A(9,10,13) 115.3201 -DE/DX = 0.0 !
! A20 A(9,10,14) 124.9751 -DE/DX = 0.0 !
! A21 A(13,10,14) 119.7048 -DE/DX = 0.0 !
! A22 A(10,14,15) 121.8623 -DE/DX = 0.0 !
! A23 A(10,14,16) 121.8382 -DE/DX = 0.0 !
! A24 A(15,14,16) 116.2993 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 64.0448 -DE/DX = 0.0 !
! D2 D(3,1,2,6) -115.9105 -DE/DX = 0.0 !
! D3 D(4,1,2,5) -178.2065 -DE/DX = 0.0 !
! D4 D(4,1,2,6) 1.8382 -DE/DX = 0.0 !
! D5 D(9,1,2,5) -56.1091 -DE/DX = 0.0 !
! D6 D(9,1,2,6) 123.9356 -DE/DX = 0.0 !
! D7 D(2,1,9,10) -64.1694 -DE/DX = 0.0 !
! D8 D(2,1,9,11) 175.1506 -DE/DX = 0.0 !
! D9 D(2,1,9,12) 58.1754 -DE/DX = 0.0 !
! D10 D(3,1,9,10) 175.1506 -DE/DX = 0.0 !
! D11 D(3,1,9,11) 54.4706 -DE/DX = 0.0 !
! D12 D(3,1,9,12) -62.5047 -DE/DX = 0.0 !
! D13 D(4,1,9,10) 58.1754 -DE/DX = 0.0 !
! D14 D(4,1,9,11) -62.5047 -DE/DX = 0.0 !
! D15 D(4,1,9,12) -179.4799 -DE/DX = 0.0 !
! D16 D(1,2,6,7) -0.3267 -DE/DX = 0.0 !
! D17 D(1,2,6,8) 179.8392 -DE/DX = 0.0 !
! D18 D(5,2,6,7) 179.7198 -DE/DX = 0.0 !
! D19 D(5,2,6,8) -0.1143 -DE/DX = 0.0 !
! D20 D(1,9,10,13) -56.1091 -DE/DX = 0.0 !
! D21 D(1,9,10,14) 123.9356 -DE/DX = 0.0 !
! D22 D(11,9,10,13) 64.0448 -DE/DX = 0.0 !
! D23 D(11,9,10,14) -115.9105 -DE/DX = 0.0 !
! D24 D(12,9,10,13) -178.2065 -DE/DX = 0.0 !
! D25 D(12,9,10,14) 1.8382 -DE/DX = 0.0 !
! D26 D(9,10,14,15) -0.3267 -DE/DX = 0.0 !
! D27 D(9,10,14,16) 179.8392 -DE/DX = 0.0 !
! D28 D(13,10,14,15) 179.7198 -DE/DX = 0.0 !
! D29 D(13,10,14,16) -0.1143 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Chair | Optimisation to a minimum | HF | 3-21G | Default | -231.69166702 a.u. | C2 |
IRC analysis of optimised boat transition state
1. Calculating minimum energy path from boat transition state
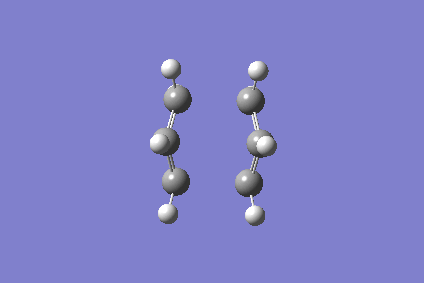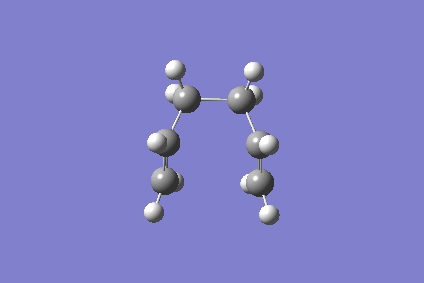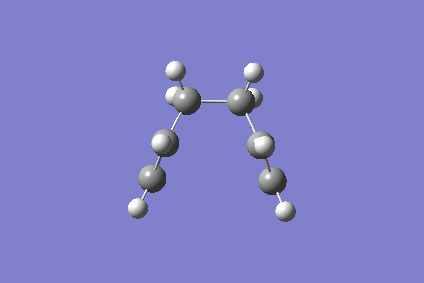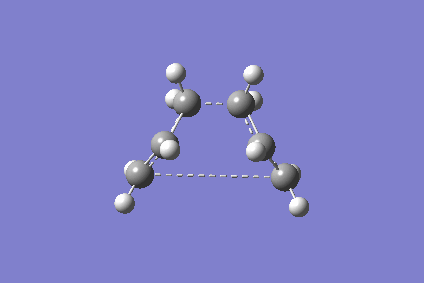
As the reaction coordinate is symmetrical in the cope rearrangement, "forward only" is chosen for this IRC calculation. There are 45 intermediate geometries obtianed, which are connected together to show the geometric change following the calculated minimum energy path from the boat transition structure to either reactant or product.
2. File link
The IRC analysed file is linked to here.
3. Structure of the last point of the IRC calculation
test molecule |
4. General information of the last point of the IRC calculation
The gradient is less than 0.001, which means the optimisation is complete.
5. Symmetry information of the last point of the IRC calculation
6. Key information of the IRC calculation
| Transition state type | Job type | Method | Basis set | Memory limit | Energy of the last point | Point group of the last point |
|---|---|---|---|---|---|---|
| Boat | IRC, forward only, calculate always, compute 50 points | HF | 3-21G | Default | -231.68298284 a.u. | Cs |
7. IRC plot of the IRC calculation
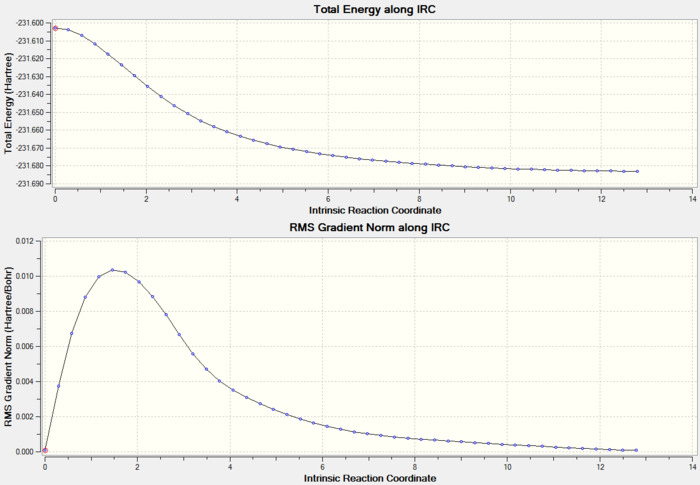
As the IRC plot is shown above, the RMS gradient is very close to 0 but not exactly equal to 0, so I decided to run a normal minimisation for the last point of this IRC calculation in order to obtain a more accurate minimum.
(1) Normal minimisation
1. Optimising last point of the IRC calculation using HF/3-21G
test molecule |
The structure looks almost the same as the one before optimisation.
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete. And the energy is the minimum I found, which is just slightly lower than that before optimisation.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000019 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000234 0.001800 YES
RMS Displacement 0.000048 0.001200 YES
Predicted change in Energy=-4.234929D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.5104 -DE/DX = 0.0 !
! R2 R(1,6) 1.5764 -DE/DX = 0.0 !
! R3 R(1,7) 1.0854 -DE/DX = 0.0 !
! R4 R(1,12) 1.0832 -DE/DX = 0.0 !
! R5 R(2,3) 1.3158 -DE/DX = 0.0 !
! R6 R(2,8) 1.0756 -DE/DX = 0.0 !
! R7 R(3,4) 4.3471 -DE/DX = 0.0 !
! R8 R(3,13) 1.0733 -DE/DX = 0.0 !
! R9 R(3,14) 1.0745 -DE/DX = 0.0 !
! R10 R(4,5) 1.3158 -DE/DX = 0.0 !
! R11 R(4,15) 1.0745 -DE/DX = 0.0 !
! R12 R(4,16) 1.0733 -DE/DX = 0.0 !
! R13 R(5,6) 1.5104 -DE/DX = 0.0 !
! R14 R(5,9) 1.0756 -DE/DX = 0.0 !
! R15 R(6,10) 1.0832 -DE/DX = 0.0 !
! R16 R(6,11) 1.0854 -DE/DX = 0.0 !
! A1 A(2,1,6) 114.7587 -DE/DX = 0.0 !
! A2 A(2,1,7) 108.6492 -DE/DX = 0.0 !
! A3 A(2,1,12) 108.7769 -DE/DX = 0.0 !
! A4 A(6,1,7) 108.6886 -DE/DX = 0.0 !
! A5 A(6,1,12) 108.6894 -DE/DX = 0.0 !
! A6 A(7,1,12) 107.0064 -DE/DX = 0.0 !
! A7 A(1,2,3) 124.4302 -DE/DX = 0.0 !
! A8 A(1,2,8) 116.122 -DE/DX = 0.0 !
! A9 A(3,2,8) 119.4388 -DE/DX = 0.0 !
! A10 A(2,3,4) 55.1012 -DE/DX = 0.0 !
! A11 A(2,3,13) 121.8398 -DE/DX = 0.0 !
! A12 A(2,3,14) 121.8492 -DE/DX = 0.0 !
! A13 A(4,3,13) 114.2083 -DE/DX = 0.0 !
! A14 A(4,3,14) 101.0171 -DE/DX = 0.0 !
! A15 A(13,3,14) 116.3106 -DE/DX = 0.0 !
! A16 A(3,4,5) 55.1012 -DE/DX = 0.0 !
! A17 A(3,4,15) 101.0171 -DE/DX = 0.0 !
! A18 A(3,4,16) 114.2083 -DE/DX = 0.0 !
! A19 A(5,4,15) 121.8492 -DE/DX = 0.0 !
! A20 A(5,4,16) 121.8398 -DE/DX = 0.0 !
! A21 A(15,4,16) 116.3106 -DE/DX = 0.0 !
! A22 A(4,5,6) 124.4302 -DE/DX = 0.0 !
! A23 A(4,5,9) 119.4388 -DE/DX = 0.0 !
! A24 A(6,5,9) 116.122 -DE/DX = 0.0 !
! A25 A(1,6,5) 114.7587 -DE/DX = 0.0 !
! A26 A(1,6,10) 108.6894 -DE/DX = 0.0 !
! A27 A(1,6,11) 108.6886 -DE/DX = 0.0 !
! A28 A(5,6,10) 108.7769 -DE/DX = 0.0 !
! A29 A(5,6,11) 108.6492 -DE/DX = 0.0 !
! A30 A(10,6,11) 107.0064 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 116.5974 -DE/DX = 0.0 !
! D2 D(6,1,2,8) -64.5 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -121.5281 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 57.3745 -DE/DX = 0.0 !
! D5 D(12,1,2,3) -5.3745 -DE/DX = 0.0 !
! D6 D(12,1,2,8) 173.5281 -DE/DX = 0.0 !
! D7 D(2,1,6,5) 0.0 -DE/DX = 0.0 !
! D8 D(2,1,6,10) -122.0194 -DE/DX = 0.0 !
! D9 D(2,1,6,11) 121.8531 -DE/DX = 0.0 !
! D10 D(7,1,6,5) -121.8531 -DE/DX = 0.0 !
! D11 D(7,1,6,10) 116.1275 -DE/DX = 0.0 !
! D12 D(7,1,6,11) 0.0 -DE/DX = 0.0 !
! D13 D(12,1,6,5) 122.0194 -DE/DX = 0.0 !
! D14 D(12,1,6,10) 0.0 -DE/DX = 0.0 !
! D15 D(12,1,6,11) -116.1275 -DE/DX = 0.0 !
! D16 D(1,2,3,4) -81.9003 -DE/DX = 0.0 !
! D17 D(1,2,3,13) 179.1634 -DE/DX = 0.0 !
! D18 D(1,2,3,14) -1.0516 -DE/DX = 0.0 !
! D19 D(8,2,3,4) 99.2311 -DE/DX = 0.0 !
! D20 D(8,2,3,13) 0.2948 -DE/DX = 0.0 !
! D21 D(8,2,3,14) -179.9202 -DE/DX = 0.0 !
! D22 D(2,3,4,5) 0.0 -DE/DX = 0.0 !
! D23 D(2,3,4,15) 121.3093 -DE/DX = 0.0 !
! D24 D(2,3,4,16) -113.0549 -DE/DX = 0.0 !
! D25 D(13,3,4,5) 113.0549 -DE/DX = 0.0 !
! D26 D(13,3,4,15) -125.6357 -DE/DX = 0.0 !
! D27 D(13,3,4,16) 0.0 -DE/DX = 0.0 !
! D28 D(14,3,4,5) -121.3093 -DE/DX = 0.0 !
! D29 D(14,3,4,15) 0.0 -DE/DX = 0.0 !
! D30 D(14,3,4,16) 125.6357 -DE/DX = 0.0 !
! D31 D(3,4,5,6) 81.9003 -DE/DX = 0.0 !
! D32 D(3,4,5,9) -99.2311 -DE/DX = 0.0 !
! D33 D(15,4,5,6) 1.0516 -DE/DX = 0.0 !
! D34 D(15,4,5,9) 179.9202 -DE/DX = 0.0 !
! D35 D(16,4,5,6) -179.1634 -DE/DX = 0.0 !
! D36 D(16,4,5,9) -0.2948 -DE/DX = 0.0 !
! D37 D(4,5,6,1) -116.5974 -DE/DX = 0.0 !
! D38 D(4,5,6,10) 5.3745 -DE/DX = 0.0 !
! D39 D(4,5,6,11) 121.5281 -DE/DX = 0.0 !
! D40 D(9,5,6,1) 64.5 -DE/DX = 0.0 !
! D41 D(9,5,6,10) -173.5281 -DE/DX = 0.0 !
! D42 D(9,5,6,11) -57.3745 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Transition state type | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Boat | Optimisation to a minimum | HF | 3-21G | Default | -231.68302549 a.u. | Cs |
(g) Reoptimisation of chair and boat transition states using B3LYP/6-31G(d)
Reoptimisation of chair transition state
1. Optimising chair transition state using B3LYP/6-31G(d)
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000027 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000107 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.303195D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4075 -DE/DX = 0.0 !
! R2 R(1,3) 1.0899 -DE/DX = 0.0 !
! R3 R(1,4) 1.0886 -DE/DX = 0.0 !
! R4 R(1,9) 1.9675 -DE/DX = 0.0 !
! R5 R(1,11) 2.4452 -DE/DX = 0.0 !
! R6 R(1,12) 2.3736 -DE/DX = 0.0 !
! R7 R(2,5) 1.0905 -DE/DX = 0.0 !
! R8 R(2,6) 1.4075 -DE/DX = 0.0 !
! R9 R(3,9) 2.4452 -DE/DX = 0.0 !
! R10 R(4,9) 2.3736 -DE/DX = 0.0 !
! R11 R(6,7) 1.0886 -DE/DX = 0.0 !
! R12 R(6,8) 1.0899 -DE/DX = 0.0 !
! R13 R(6,14) 1.9675 -DE/DX = 0.0 !
! R14 R(6,15) 2.3736 -DE/DX = 0.0 !
! R15 R(6,16) 2.4452 -DE/DX = 0.0 !
! R16 R(7,14) 2.3736 -DE/DX = 0.0 !
! R17 R(8,14) 2.4452 -DE/DX = 0.0 !
! R18 R(9,10) 1.4075 -DE/DX = 0.0 !
! R19 R(9,11) 1.0899 -DE/DX = 0.0 !
! R20 R(9,12) 1.0886 -DE/DX = 0.0 !
! R21 R(10,13) 1.0905 -DE/DX = 0.0 !
! R22 R(10,14) 1.4075 -DE/DX = 0.0 !
! R23 R(14,15) 1.0886 -DE/DX = 0.0 !
! R24 R(14,16) 1.0899 -DE/DX = 0.0 !
! A1 A(2,1,3) 118.2534 -DE/DX = 0.0 !
! A2 A(2,1,4) 117.9645 -DE/DX = 0.0 !
! A3 A(2,1,9) 103.6352 -DE/DX = 0.0 !
! A4 A(2,1,11) 129.4131 -DE/DX = 0.0 !
! A5 A(2,1,12) 92.2714 -DE/DX = 0.0 !
! A6 A(3,1,4) 112.4945 -DE/DX = 0.0 !
! A7 A(3,1,11) 88.5074 -DE/DX = 0.0 !
! A8 A(3,1,12) 86.7983 -DE/DX = 0.0 !
! A9 A(4,1,11) 83.1889 -DE/DX = 0.0 !
! A10 A(4,1,12) 124.4235 -DE/DX = 0.0 !
! A11 A(11,1,12) 44.126 -DE/DX = 0.0 !
! A12 A(1,2,5) 117.6354 -DE/DX = 0.0 !
! A13 A(1,2,6) 119.951 -DE/DX = 0.0 !
! A14 A(5,2,6) 117.6354 -DE/DX = 0.0 !
! A15 A(2,6,7) 117.9645 -DE/DX = 0.0 !
! A16 A(2,6,8) 118.2534 -DE/DX = 0.0 !
! A17 A(2,6,14) 103.6353 -DE/DX = 0.0 !
! A18 A(2,6,15) 92.2714 -DE/DX = 0.0 !
! A19 A(2,6,16) 129.4132 -DE/DX = 0.0 !
! A20 A(7,6,8) 112.4944 -DE/DX = 0.0 !
! A21 A(7,6,15) 124.4236 -DE/DX = 0.0 !
! A22 A(7,6,16) 83.189 -DE/DX = 0.0 !
! A23 A(8,6,15) 86.7983 -DE/DX = 0.0 !
! A24 A(8,6,16) 88.5075 -DE/DX = 0.0 !
! A25 A(15,6,16) 44.126 -DE/DX = 0.0 !
! A26 A(1,9,10) 103.6352 -DE/DX = 0.0 !
! A27 A(3,9,4) 44.126 -DE/DX = 0.0 !
! A28 A(3,9,10) 129.4132 -DE/DX = 0.0 !
! A29 A(3,9,11) 88.5075 -DE/DX = 0.0 !
! A30 A(3,9,12) 83.1889 -DE/DX = 0.0 !
! A31 A(4,9,10) 92.2714 -DE/DX = 0.0 !
! A32 A(4,9,11) 86.7983 -DE/DX = 0.0 !
! A33 A(4,9,12) 124.4236 -DE/DX = 0.0 !
! A34 A(10,9,11) 118.2534 -DE/DX = 0.0 !
! A35 A(10,9,12) 117.9645 -DE/DX = 0.0 !
! A36 A(11,9,12) 112.4945 -DE/DX = 0.0 !
! A37 A(9,10,13) 117.6354 -DE/DX = 0.0 !
! A38 A(9,10,14) 119.951 -DE/DX = 0.0 !
! A39 A(13,10,14) 117.6354 -DE/DX = 0.0 !
! A40 A(6,14,10) 103.6352 -DE/DX = 0.0 !
! A41 A(7,14,8) 44.126 -DE/DX = 0.0 !
! A42 A(7,14,10) 92.2714 -DE/DX = 0.0 !
! A43 A(7,14,15) 124.4236 -DE/DX = 0.0 !
! A44 A(7,14,16) 86.7983 -DE/DX = 0.0 !
! A45 A(8,14,10) 129.4132 -DE/DX = 0.0 !
! A46 A(8,14,15) 83.1889 -DE/DX = 0.0 !
! A47 A(8,14,16) 88.5074 -DE/DX = 0.0 !
! A48 A(10,14,15) 117.9645 -DE/DX = 0.0 !
! A49 A(10,14,16) 118.2534 -DE/DX = 0.0 !
! A50 A(15,14,16) 112.4944 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 22.6226 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 177.5758 -DE/DX = 0.0 !
! D3 D(4,1,2,5) 163.6158 -DE/DX = 0.0 !
! D4 D(4,1,2,6) -41.431 -DE/DX = 0.0 !
! D5 D(9,1,2,5) -89.7714 -DE/DX = 0.0 !
! D6 D(9,1,2,6) 65.1818 -DE/DX = 0.0 !
! D7 D(11,1,2,5) -91.1655 -DE/DX = 0.0 !
! D8 D(11,1,2,6) 63.7877 -DE/DX = 0.0 !
! D9 D(12,1,2,5) -64.9622 -DE/DX = 0.0 !
! D10 D(12,1,2,6) 89.991 -DE/DX = 0.0 !
! D11 D(2,1,9,10) -54.0215 -DE/DX = 0.0 !
! D12 D(1,2,6,7) 41.4311 -DE/DX = 0.0 !
! D13 D(1,2,6,8) -177.5759 -DE/DX = 0.0 !
! D14 D(1,2,6,14) -65.1818 -DE/DX = 0.0 !
! D15 D(1,2,6,15) -89.991 -DE/DX = 0.0 !
! D16 D(1,2,6,16) -63.7878 -DE/DX = 0.0 !
! D17 D(5,2,6,7) -163.6157 -DE/DX = 0.0 !
! D18 D(5,2,6,8) -22.6227 -DE/DX = 0.0 !
! D19 D(5,2,6,14) 89.7714 -DE/DX = 0.0 !
! D20 D(5,2,6,15) 64.9622 -DE/DX = 0.0 !
! D21 D(5,2,6,16) 91.1655 -DE/DX = 0.0 !
! D22 D(2,6,14,10) 54.0216 -DE/DX = 0.0 !
! D23 D(1,9,10,13) -89.7714 -DE/DX = 0.0 !
! D24 D(1,9,10,14) 65.1818 -DE/DX = 0.0 !
! D25 D(3,9,10,13) -91.1655 -DE/DX = 0.0 !
! D26 D(3,9,10,14) 63.7877 -DE/DX = 0.0 !
! D27 D(4,9,10,13) -64.9622 -DE/DX = 0.0 !
! D28 D(4,9,10,14) 89.991 -DE/DX = 0.0 !
! D29 D(11,9,10,13) 22.6227 -DE/DX = 0.0 !
! D30 D(11,9,10,14) 177.5759 -DE/DX = 0.0 !
! D31 D(12,9,10,13) 163.6158 -DE/DX = 0.0 !
! D32 D(12,9,10,14) -41.431 -DE/DX = 0.0 !
! D33 D(9,10,14,6) -65.1818 -DE/DX = 0.0 !
! D34 D(9,10,14,7) -89.991 -DE/DX = 0.0 !
! D35 D(9,10,14,8) -63.7877 -DE/DX = 0.0 !
! D36 D(9,10,14,15) 41.431 -DE/DX = 0.0 !
! D37 D(9,10,14,16) -177.5758 -DE/DX = 0.0 !
! D38 D(13,10,14,6) 89.7714 -DE/DX = 0.0 !
! D39 D(13,10,14,7) 64.9622 -DE/DX = 0.0 !
! D40 D(13,10,14,8) 91.1655 -DE/DX = 0.0 !
! D41 D(13,10,14,15) -163.6158 -DE/DX = 0.0 !
! D42 D(13,10,14,16) -22.6226 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
The point group of the optimised structure is C2h, confirming the structure is correct.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 565.53 cm-1 which is lower than that of the HF/3-21G optimised structure (i.e. 818.02 cm-1). The corresponding vibrational mode is shown below, and the motion is similar to that of the HF/3-21G optimised structure but slower.
7. Thermochemistry
Sum of electronic and zero-point Energies= -234.414929 Sum of electronic and thermal Energies= -234.409008 Sum of electronic and thermal Enthalpies= -234.408064 Sum of electronic and thermal Free Energies= -234.443814
8. Key information
| Transition state type | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Chair | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | B3LYP | 6-31G(d) | Default | -234.55698303 a.u. | C2h |
Reoptimisation of boat transition state
1. Optimising boat transition state using B3LYP/6-31G(d)
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000129 0.000450 YES
RMS Force 0.000025 0.000300 YES
Maximum Displacement 0.001795 0.001800 YES
RMS Displacement 0.000585 0.001200 YES
Predicted change in Energy=-1.407564D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3931 -DE/DX = 0.0 !
! R2 R(1,6) 2.2073 -DE/DX = 0.0001 !
! R3 R(1,7) 1.087 -DE/DX = 0.0 !
! R4 R(1,12) 1.0868 -DE/DX = 0.0 !
! R5 R(2,3) 1.3934 -DE/DX = 0.0 !
! R6 R(2,8) 1.0911 -DE/DX = 0.0 !
! R7 R(3,4) 2.2053 -DE/DX = 0.0 !
! R8 R(3,13) 1.087 -DE/DX = 0.0 !
! R9 R(3,14) 1.0868 -DE/DX = 0.0 !
! R10 R(4,5) 1.3934 -DE/DX = 0.0 !
! R11 R(4,15) 1.0868 -DE/DX = 0.0 !
! R12 R(4,16) 1.087 -DE/DX = 0.0 !
! R13 R(5,6) 1.3931 -DE/DX = 0.0 !
! R14 R(5,9) 1.0911 -DE/DX = 0.0 !
! R15 R(6,10) 1.0868 -DE/DX = 0.0 !
! R16 R(6,11) 1.087 -DE/DX = 0.0 !
! A1 A(2,1,6) 103.479 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.7469 -DE/DX = 0.0 !
! A3 A(2,1,12) 118.9444 -DE/DX = 0.0 !
! A4 A(6,1,7) 101.9234 -DE/DX = 0.0 !
! A5 A(6,1,12) 90.4694 -DE/DX = 0.0 !
! A6 A(7,1,12) 114.4414 -DE/DX = 0.0 !
! A7 A(1,2,3) 122.2408 -DE/DX = 0.0001 !
! A8 A(1,2,8) 117.1548 -DE/DX = 0.0 !
! A9 A(3,2,8) 117.1587 -DE/DX = 0.0 !
! A10 A(2,3,4) 103.5173 -DE/DX = 0.0 !
! A11 A(2,3,13) 119.7332 -DE/DX = 0.0 !
! A12 A(2,3,14) 118.9246 -DE/DX = 0.0 !
! A13 A(4,3,13) 101.9344 -DE/DX = 0.0 !
! A14 A(4,3,14) 90.4988 -DE/DX = 0.0 !
! A15 A(13,3,14) 114.4342 -DE/DX = 0.0 !
! A16 A(3,4,5) 103.5173 -DE/DX = 0.0 !
! A17 A(3,4,15) 90.4988 -DE/DX = 0.0 !
! A18 A(3,4,16) 101.9344 -DE/DX = 0.0 !
! A19 A(5,4,15) 118.9246 -DE/DX = 0.0 !
! A20 A(5,4,16) 119.7332 -DE/DX = 0.0 !
! A21 A(15,4,16) 114.4342 -DE/DX = 0.0 !
! A22 A(4,5,6) 122.2408 -DE/DX = 0.0001 !
! A23 A(4,5,9) 117.1587 -DE/DX = 0.0 !
! A24 A(6,5,9) 117.1548 -DE/DX = 0.0 !
! A25 A(1,6,5) 103.479 -DE/DX = 0.0 !
! A26 A(1,6,10) 90.4694 -DE/DX = 0.0 !
! A27 A(1,6,11) 101.9234 -DE/DX = 0.0 !
! A28 A(5,6,10) 118.9444 -DE/DX = 0.0 !
! A29 A(5,6,11) 119.7469 -DE/DX = 0.0 !
! A30 A(10,6,11) 114.4414 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 64.1921 -DE/DX = 0.0 !
! D2 D(6,1,2,8) -94.2493 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 176.6297 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 18.1883 -DE/DX = 0.0 !
! D5 D(12,1,2,3) -33.9823 -DE/DX = 0.0 !
! D6 D(12,1,2,8) 167.5763 -DE/DX = 0.0 !
! D7 D(2,1,6,5) 0.0 -DE/DX = 0.0 !
! D8 D(2,1,6,10) -119.9768 -DE/DX = 0.0 !
! D9 D(2,1,6,11) 124.8959 -DE/DX = 0.0 !
! D10 D(7,1,6,5) -124.8959 -DE/DX = 0.0 !
! D11 D(7,1,6,10) 115.1273 -DE/DX = 0.0 !
! D12 D(7,1,6,11) 0.0 -DE/DX = 0.0 !
! D13 D(12,1,6,5) 119.9768 -DE/DX = 0.0 !
! D14 D(12,1,6,10) 0.0 -DE/DX = 0.0 !
! D15 D(12,1,6,11) -115.1273 -DE/DX = 0.0 !
! D16 D(1,2,3,4) -64.2111 -DE/DX = 0.0 !
! D17 D(1,2,3,13) -176.6833 -DE/DX = 0.0 !
! D18 D(1,2,3,14) 34.0145 -DE/DX = 0.0 !
! D19 D(8,2,3,4) 94.2295 -DE/DX = 0.0 !
! D20 D(8,2,3,13) -18.2427 -DE/DX = 0.0 !
! D21 D(8,2,3,14) -167.5449 -DE/DX = 0.0 !
! D22 D(2,3,4,5) 0.0 -DE/DX = 0.0 !
! D23 D(2,3,4,15) 119.9702 -DE/DX = 0.0 !
! D24 D(2,3,4,16) -124.9018 -DE/DX = 0.0 !
! D25 D(13,3,4,5) 124.9018 -DE/DX = 0.0 !
! D26 D(13,3,4,15) -115.128 -DE/DX = 0.0 !
! D27 D(13,3,4,16) 0.0 -DE/DX = 0.0 !
! D28 D(14,3,4,5) -119.9702 -DE/DX = 0.0 !
! D29 D(14,3,4,15) 0.0 -DE/DX = 0.0 !
! D30 D(14,3,4,16) 115.128 -DE/DX = 0.0 !
! D31 D(3,4,5,6) 64.2111 -DE/DX = 0.0 !
! D32 D(3,4,5,9) -94.2295 -DE/DX = 0.0 !
! D33 D(15,4,5,6) -34.0145 -DE/DX = 0.0 !
! D34 D(15,4,5,9) 167.5449 -DE/DX = 0.0 !
! D35 D(16,4,5,6) 176.6833 -DE/DX = 0.0 !
! D36 D(16,4,5,9) 18.2427 -DE/DX = 0.0 !
! D37 D(4,5,6,1) -64.1921 -DE/DX = 0.0 !
! D38 D(4,5,6,10) 33.9823 -DE/DX = 0.0 !
! D39 D(4,5,6,11) -176.6297 -DE/DX = 0.0 !
! D40 D(9,5,6,1) 94.2493 -DE/DX = 0.0 !
! D41 D(9,5,6,10) -167.5763 -DE/DX = 0.0 !
! D42 D(9,5,6,11) -18.1883 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
The point group of the optimised structure is C2v, confirming the structure is correct.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 530.43 cm-1 which is lower than that of the HF/3-21G optimised structure (i.e. 839.62 cm-1). The corresponding vibrational mode is shown below, and the motion is similar to that of the HF/3-21G optimised structure but slower.
7. Thermochemistry
Sum of electronic and zero-point Energies= -234.402336 Sum of electronic and thermal Energies= -234.396001 Sum of electronic and thermal Enthalpies= -234.395057 Sum of electronic and thermal Free Energies= -234.431746
8. Key information
| Transition state type | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Boat | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | B3LYP | 6-31G(d) | Default | -231.54309298 a.u. | C2v |
Result and discussion
1. Geometric data comparison
(1) Different optimisations of chair transition state
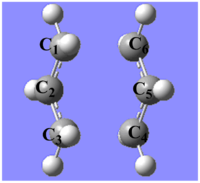
| Geometric parameter | HF/3-21G | B3LYP/6-31G(d) |
|---|---|---|
| C1=C2 (or C2=C3, C4=C5, C5=C6) bond length | 1.38928 Å | 1.40749 Å |
| C3-C4 (or C1=C6) bond length | 2.02038 Å | 1.96751 Å |
| C2-C5 bond length | 2.87892 Å | 2.90970 Å |
| C2=C3-C4-C5 (or C2-C1-C6=C5) dihedral angle | +(or-)54.958o | +(or-)54.022o |
The difference in geometric data is small for using HF/3-21G and B3LYP/6-31G(d) as the method/basis set for optimisation of chair transition state.
(2) Different optimisations of boat transition state
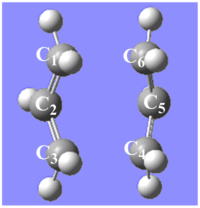
| Geometric parameter | HF/3-21G | B3LYP/6-31G(d) |
|---|---|---|
| C1=C2 (or C2=C3, C4=C5, C5=C6) bond length | 1.38154 Å | 1.39307 Å |
| C3-C4 (or C1=C6) bond length | 2.14182 Å | 2.20727 Å |
| C2-C5 bond length | 2.77903 Å | 2.85669 Å |
| C2=C3-C4-C5 (or C2-C1-C6=C5) dihedral angle | 0o | 0o |
Similarly, the difference in geometric data is small for using HF/3-21G and B3LYP/6-31G(d) as the method/basis set for optimisation of boat transition state.
2. Thermochemistry data comparison
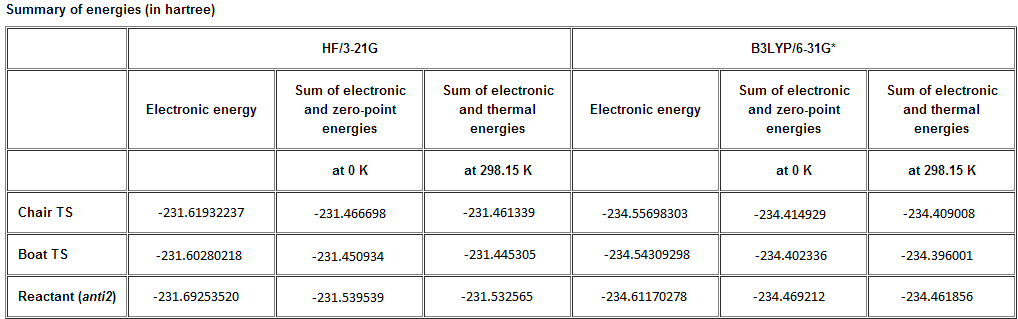
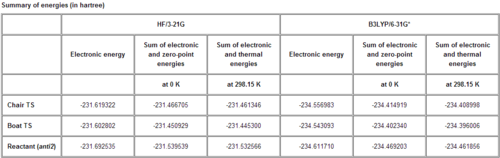
As the energy table shown above, the energy difference between optimisations using HF/3-21G and B3LYP/6-31G(d) is huge, which is approximately 3 a.u. (or 1883 kcal/mol). Moreover, my experimental data is consistent with the data given in literature which is shown on the right.
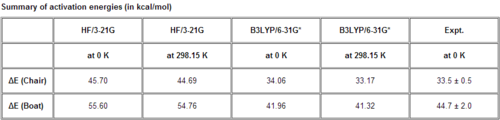
My calculated activation energy data also gives a good agreement to the data given in literature which is shown on the right.
The Diels Alder Cycloaddition
Diels Alder Reaction Between Cis-Butadiene and Ethylene
Optimising the Reactants
(a) Optimisation of cis-butadiene
1. Optimising cis butadiene using AM1 method
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000051 0.000300 YES
Maximum Displacement 0.000767 0.001800 YES
RMS Displacement 0.000254 0.001200 YES
Predicted change in Energy=-1.540655D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.335 -DE/DX = 0.0001 !
! R2 R(1,3) 1.0979 -DE/DX = -0.0001 !
! R3 R(1,4) 1.0977 -DE/DX = 0.0 !
! R4 R(2,5) 1.1055 -DE/DX = -0.0002 !
! R5 R(2,6) 1.4495 -DE/DX = 0.0 !
! R6 R(6,7) 1.335 -DE/DX = 0.0001 !
! R7 R(6,8) 1.1055 -DE/DX = -0.0002 !
! R8 R(7,9) 1.0979 -DE/DX = -0.0001 !
! R9 R(7,10) 1.0977 -DE/DX = 0.0 !
! A1 A(2,1,3) 121.928 -DE/DX = 0.0 !
! A2 A(2,1,4) 123.1583 -DE/DX = 0.0 !
! A3 A(3,1,4) 114.9137 -DE/DX = 0.0001 !
! A4 A(1,2,5) 119.8212 -DE/DX = 0.0 !
! A5 A(1,2,6) 125.6618 -DE/DX = 0.0 !
! A6 A(5,2,6) 114.5169 -DE/DX = 0.0 !
! A7 A(2,6,7) 125.6618 -DE/DX = 0.0 !
! A8 A(2,6,8) 114.5169 -DE/DX = 0.0 !
! A9 A(7,6,8) 119.8212 -DE/DX = 0.0 !
! A10 A(6,7,9) 121.928 -DE/DX = 0.0 !
! A11 A(6,7,10) 123.1583 -DE/DX = 0.0 !
! A12 A(9,7,10) 114.9137 -DE/DX = 0.0001 !
! D1 D(3,1,2,5) 0.0 -DE/DX = 0.0 !
! D2 D(3,1,2,6) -180.0003 -DE/DX = 0.0 !
! D3 D(4,1,2,5) 180.0002 -DE/DX = 0.0 !
! D4 D(4,1,2,6) 0.0 -DE/DX = 0.0 !
! D5 D(1,2,6,7) 0.0004 -DE/DX = 0.0 !
! D6 D(1,2,6,8) -180.0001 -DE/DX = 0.0 !
! D7 D(5,2,6,7) -179.9998 -DE/DX = 0.0 !
! D8 D(5,2,6,8) -0.0003 -DE/DX = 0.0 !
! D9 D(2,6,7,9) -180.0005 -DE/DX = 0.0 !
! D10 D(2,6,7,10) -0.0001 -DE/DX = 0.0 !
! D11 D(8,6,7,9) 0.0001 -DE/DX = 0.0 !
! D12 D(8,6,7,10) 180.0005 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
7. Key information
| Molecule | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Cis-butadiene | Optimisation to a minimum | Semi-empirical molecular orbital, AM1 | ZDO | Default | 0.04879734 a.u. | C2v |
(b) Optimisation of ethylene
1. Optimising ethylene using AM1 method
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000031 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000057 0.001800 YES
RMS Displacement 0.000037 0.001200 YES
Predicted change in Energy=-2.644693D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3259 -DE/DX = 0.0 !
! R2 R(1,3) 1.0983 -DE/DX = 0.0 !
! R3 R(1,4) 1.0983 -DE/DX = 0.0 !
! R4 R(2,5) 1.0983 -DE/DX = 0.0 !
! R5 R(2,6) 1.0983 -DE/DX = 0.0 !
! A1 A(2,1,3) 122.7159 -DE/DX = 0.0 !
! A2 A(2,1,4) 122.718 -DE/DX = 0.0 !
! A3 A(3,1,4) 114.5661 -DE/DX = 0.0 !
! A4 A(1,2,5) 122.7159 -DE/DX = 0.0 !
! A5 A(1,2,6) 122.718 -DE/DX = 0.0 !
! A6 A(5,2,6) 114.5661 -DE/DX = 0.0 !
! D1 D(3,1,2,5) 180.0012 -DE/DX = 0.0 !
! D2 D(3,1,2,6) 0.0016 -DE/DX = 0.0 !
! D3 D(4,1,2,5) -0.0002 -DE/DX = 0.0 !
! D4 D(4,1,2,6) -179.9998 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Molecule | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Ethylene | Optimisation to a minimum | Semi-empirical molecular orbital, AM1 | ZDO | Default | 0.02619024 a.u. | D2h |
Optimising the Transition Structure
(a) Optimisation of guess transition state
1. Optimising guess transition state using AM1 method
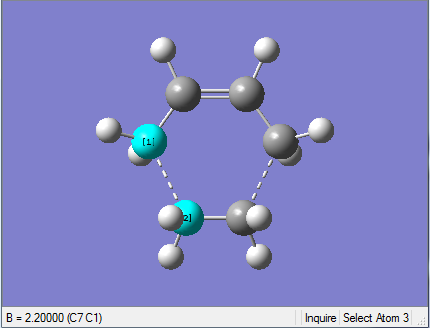
The guess transition state was drawn as above by combining the optimised ethylene and butadiene structures with two partially formed C-C bonds of 2.2 Å bond length and modifying the H-C-H bond angles. The optimised structure is shown below, which has 2.11929 Å bond lengths for the partially formed bonds.
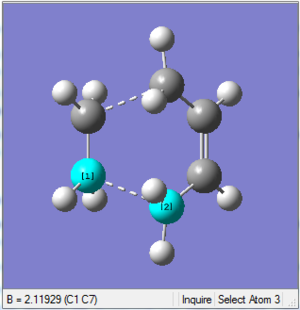
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000023 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000139 0.001800 YES
RMS Displacement 0.000042 0.001200 YES
Predicted change in Energy=-4.471972D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3829 -DE/DX = 0.0 !
! R2 R(1,7) 2.1193 -DE/DX = 0.0 !
! R3 R(1,15) 1.0996 -DE/DX = 0.0 !
! R4 R(1,16) 1.1002 -DE/DX = 0.0 !
! R5 R(2,3) 2.1192 -DE/DX = 0.0 !
! R6 R(2,13) 1.0996 -DE/DX = 0.0 !
! R7 R(2,14) 1.1002 -DE/DX = 0.0 !
! R8 R(3,4) 1.3818 -DE/DX = 0.0 !
! R9 R(3,11) 1.1008 -DE/DX = 0.0 !
! R10 R(3,12) 1.0989 -DE/DX = 0.0 !
! R11 R(4,5) 1.1018 -DE/DX = 0.0 !
! R12 R(4,6) 1.3975 -DE/DX = 0.0 !
! R13 R(6,7) 1.3819 -DE/DX = 0.0 !
! R14 R(6,8) 1.1018 -DE/DX = 0.0 !
! R15 R(7,9) 1.0989 -DE/DX = 0.0 !
! R16 R(7,10) 1.1008 -DE/DX = 0.0 !
! A1 A(2,1,7) 109.9397 -DE/DX = 0.0 !
! A2 A(2,1,15) 120.0126 -DE/DX = 0.0 !
! A3 A(2,1,16) 119.9894 -DE/DX = 0.0 !
! A4 A(7,1,15) 90.8574 -DE/DX = 0.0 !
! A5 A(7,1,16) 90.1772 -DE/DX = 0.0 !
! A6 A(15,1,16) 115.2775 -DE/DX = 0.0 !
! A7 A(1,2,3) 109.941 -DE/DX = 0.0 !
! A8 A(1,2,13) 120.0109 -DE/DX = 0.0 !
! A9 A(1,2,14) 119.9897 -DE/DX = 0.0 !
! A10 A(3,2,13) 90.8556 -DE/DX = 0.0 !
! A11 A(3,2,14) 90.1806 -DE/DX = 0.0 !
! A12 A(13,2,14) 115.2778 -DE/DX = 0.0 !
! A13 A(2,3,4) 99.3392 -DE/DX = 0.0 !
! A14 A(2,3,11) 88.8726 -DE/DX = 0.0 !
! A15 A(2,3,12) 101.637 -DE/DX = 0.0 !
! A16 A(4,3,11) 121.2502 -DE/DX = 0.0 !
! A17 A(4,3,12) 119.9973 -DE/DX = 0.0 !
! A18 A(11,3,12) 114.7406 -DE/DX = 0.0 !
! A19 A(3,4,5) 119.648 -DE/DX = 0.0 !
! A20 A(3,4,6) 121.1811 -DE/DX = 0.0 !
! A21 A(5,4,6) 118.3949 -DE/DX = 0.0 !
! A22 A(4,6,7) 121.185 -DE/DX = 0.0 !
! A23 A(4,6,8) 118.3929 -DE/DX = 0.0 !
! A24 A(7,6,8) 119.6444 -DE/DX = 0.0 !
! A25 A(1,7,6) 99.339 -DE/DX = 0.0 !
! A26 A(1,7,9) 101.6423 -DE/DX = 0.0 !
! A27 A(1,7,10) 88.868 -DE/DX = 0.0 !
! A28 A(6,7,9) 119.9974 -DE/DX = 0.0 !
! A29 A(6,7,10) 121.247 -DE/DX = 0.0 !
! A30 A(9,7,10) 114.7435 -DE/DX = 0.0 !
! D1 D(7,1,2,3) 0.0002 -DE/DX = 0.0 !
! D2 D(7,1,2,13) 103.1737 -DE/DX = 0.0 !
! D3 D(7,1,2,14) -102.3125 -DE/DX = 0.0 !
! D4 D(15,1,2,3) -103.1756 -DE/DX = 0.0 !
! D5 D(15,1,2,13) -0.002 -DE/DX = 0.0 !
! D6 D(15,1,2,14) 154.5117 -DE/DX = 0.0 !
! D7 D(16,1,2,3) 102.3078 -DE/DX = 0.0 !
! D8 D(16,1,2,13) -154.5186 -DE/DX = 0.0 !
! D9 D(16,1,2,14) -0.0049 -DE/DX = 0.0 !
! D10 D(2,1,7,6) -51.8386 -DE/DX = 0.0 !
! D11 D(2,1,7,9) -175.2883 -DE/DX = 0.0 !
! D12 D(2,1,7,10) 69.6647 -DE/DX = 0.0 !
! D13 D(15,1,7,6) 70.6802 -DE/DX = 0.0 !
! D14 D(15,1,7,9) -52.7695 -DE/DX = 0.0 !
! D15 D(15,1,7,10) -167.8164 -DE/DX = 0.0 !
! D16 D(16,1,7,6) -174.0362 -DE/DX = 0.0 !
! D17 D(16,1,7,9) 62.5142 -DE/DX = 0.0 !
! D18 D(16,1,7,10) -52.5328 -DE/DX = 0.0 !
! D19 D(1,2,3,4) 51.8417 -DE/DX = 0.0 !
! D20 D(1,2,3,11) -69.6659 -DE/DX = 0.0 !
! D21 D(1,2,3,12) 175.2895 -DE/DX = 0.0 !
! D22 D(13,2,3,4) -70.6748 -DE/DX = 0.0 !
! D23 D(13,2,3,11) 167.8176 -DE/DX = 0.0 !
! D24 D(13,2,3,12) 52.773 -DE/DX = 0.0 !
! D25 D(14,2,3,4) 174.0412 -DE/DX = 0.0 !
! D26 D(14,2,3,11) 52.5336 -DE/DX = 0.0 !
! D27 D(14,2,3,12) -62.511 -DE/DX = 0.0 !
! D28 D(2,3,4,5) 109.9775 -DE/DX = 0.0 !
! D29 D(2,3,4,6) -59.7708 -DE/DX = 0.0 !
! D30 D(11,3,4,5) -155.6367 -DE/DX = 0.0 !
! D31 D(11,3,4,6) 34.615 -DE/DX = 0.0 !
! D32 D(12,3,4,5) 0.6493 -DE/DX = 0.0 !
! D33 D(12,3,4,6) -169.099 -DE/DX = 0.0 !
! D34 D(3,4,6,7) 0.0055 -DE/DX = 0.0 !
! D35 D(3,4,6,8) 169.8677 -DE/DX = 0.0 !
! D36 D(5,4,6,7) -169.8678 -DE/DX = 0.0 !
! D37 D(5,4,6,8) -0.0055 -DE/DX = 0.0 !
! D38 D(4,6,7,1) 59.7623 -DE/DX = 0.0 !
! D39 D(4,6,7,9) 169.0967 -DE/DX = 0.0 !
! D40 D(4,6,7,10) -34.6173 -DE/DX = 0.0 !
! D41 D(8,6,7,1) -109.975 -DE/DX = 0.0 !
! D42 D(8,6,7,9) -0.6406 -DE/DX = 0.0 !
! D43 D(8,6,7,10) 155.6454 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 956.25 cm-1. The corresponding vibrational mode is shown below, in which the C atoms of ethylene and the two terminal C atoms of butadiene approach each other in a sychronised motion and facilitates two simultaneous C-C bonds formation. This is exactly what happens in the [4+2] cycloaddtion, that is, two or more C-C bonds form at the same time (i.e. a concerted process) in a 6-membered ring like system.
7. Thermochemistry
Sum of electronic and zero-point Energies= 0.253276 Sum of electronic and thermal Energies= 0.259453 Sum of electronic and thermal Enthalpies= 0.260397 Sum of electronic and thermal Free Energies= 0.224016
9. Key information
| Subject | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Transition state | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | Semi-empirical molecular orbital, AM1 | ZDO | Default | 0.11165465 a.u. | Cs |
(b) IRC analysis of optimised transition state
1. Calculating minimum energy path from transition state
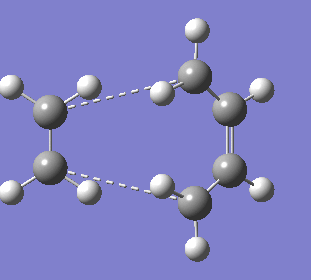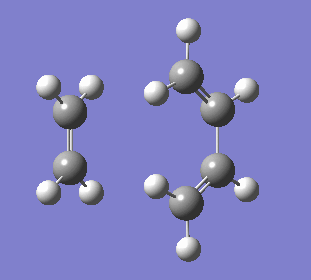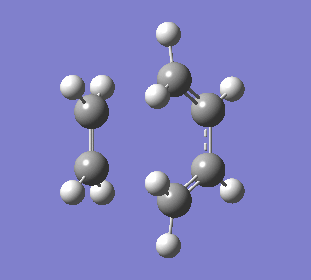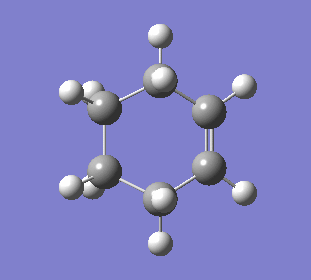
As the reaction coordinate is not symmetrical in the Diels Alder cycloaddition, "both directions" is chosen for this IRC calculation. There are 87 intermediate geometries, which are connected together to show the geometric change following the calculated minimum energy path from reactant to product via the transition state. The structure of the last point of this IRC calculation is shown below.
test molecule |
2. File link
The IRC analysed file is linked to here.
As the IRC plot is shown above, the energy minimum is reached in this calculation because the RMS gradient reaches 0 in the end. Therefore no need to conduct further calculation. The general and symmetry information of the last point of this IRC calculation is given in the following.
The gradient is less than 0.001, which means the optimisation is complete.
6. Key information
| Subject | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Transition state | IRC, both directions, calculate always, compute 100 points | Semi-empirical molecular orbital, AM1 | ZDO | Default | -0.01099223 a.u. | Cs |
(c) Reoptimisation of transition state using B3LYP/6-31G(d)
1. Optimising transition state using B3LYP/6-31G(d)
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000013 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000496 0.001800 YES
RMS Displacement 0.000106 0.001200 YES
Predicted change in Energy=-7.705534D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.386 -DE/DX = 0.0 !
! R2 R(1,6) 2.9004 -DE/DX = 0.0 !
! R3 R(1,7) 2.2725 -DE/DX = 0.0 !
! R4 R(1,9) 2.7536 -DE/DX = 0.0 !
! R5 R(1,10) 2.4251 -DE/DX = 0.0 !
! R6 R(1,15) 1.0842 -DE/DX = 0.0 !
! R7 R(1,16) 1.0863 -DE/DX = 0.0 !
! R8 R(2,3) 2.2725 -DE/DX = 0.0 !
! R9 R(2,4) 2.9004 -DE/DX = 0.0 !
! R10 R(2,11) 2.4251 -DE/DX = 0.0 !
! R11 R(2,12) 2.7536 -DE/DX = 0.0 !
! R12 R(2,13) 1.0842 -DE/DX = 0.0 !
! R13 R(2,14) 1.0863 -DE/DX = 0.0 !
! R14 R(3,4) 1.383 -DE/DX = 0.0 !
! R15 R(3,11) 1.084 -DE/DX = 0.0 !
! R16 R(3,12) 1.0873 -DE/DX = 0.0 !
! R17 R(3,13) 2.5399 -DE/DX = 0.0 !
! R18 R(3,14) 2.5323 -DE/DX = 0.0 !
! R19 R(4,5) 1.0891 -DE/DX = 0.0 !
! R20 R(4,6) 1.4072 -DE/DX = 0.0 !
! R21 R(6,7) 1.383 -DE/DX = 0.0 !
! R22 R(6,8) 1.0891 -DE/DX = 0.0 !
! R23 R(7,9) 1.0873 -DE/DX = 0.0 !
! R24 R(7,10) 1.084 -DE/DX = 0.0 !
! R25 R(7,15) 2.5399 -DE/DX = 0.0 !
! R26 R(7,16) 2.5323 -DE/DX = 0.0 !
! A1 A(2,1,6) 90.2098 -DE/DX = 0.0 !
! A2 A(2,1,7) 109.1164 -DE/DX = 0.0 !
! A3 A(2,1,9) 131.4998 -DE/DX = 0.0 !
! A4 A(2,1,10) 98.9089 -DE/DX = 0.0 !
! A5 A(2,1,15) 120.0522 -DE/DX = 0.0 !
! A6 A(2,1,16) 119.9852 -DE/DX = 0.0 !
! A7 A(6,1,9) 44.4899 -DE/DX = 0.0 !
! A8 A(6,1,10) 46.5393 -DE/DX = 0.0 !
! A9 A(6,1,15) 83.3596 -DE/DX = 0.0 !
! A10 A(6,1,16) 118.2474 -DE/DX = 0.0 !
! A11 A(9,1,10) 40.6754 -DE/DX = 0.0 !
! A12 A(9,1,15) 77.8409 -DE/DX = 0.0 !
! A13 A(9,1,16) 80.5628 -DE/DX = 0.0 !
! A14 A(10,1,15) 116.9474 -DE/DX = 0.0 !
! A15 A(10,1,16) 74.6439 -DE/DX = 0.0 !
! A16 A(15,1,16) 115.1638 -DE/DX = 0.0 !
! A17 A(1,2,3) 109.1165 -DE/DX = 0.0 !
! A18 A(1,2,4) 90.2099 -DE/DX = 0.0 !
! A19 A(1,2,11) 98.9089 -DE/DX = 0.0 !
! A20 A(1,2,12) 131.5 -DE/DX = 0.0 !
! A21 A(1,2,13) 120.0522 -DE/DX = 0.0 !
! A22 A(1,2,14) 119.9852 -DE/DX = 0.0 !
! A23 A(4,2,11) 46.5393 -DE/DX = 0.0 !
! A24 A(4,2,12) 44.49 -DE/DX = 0.0 !
! A25 A(4,2,13) 83.3596 -DE/DX = 0.0 !
! A26 A(4,2,14) 118.2474 -DE/DX = 0.0 !
! A27 A(11,2,12) 40.6755 -DE/DX = 0.0 !
! A28 A(11,2,13) 116.9475 -DE/DX = 0.0 !
! A29 A(11,2,14) 74.644 -DE/DX = 0.0 !
! A30 A(12,2,13) 77.8409 -DE/DX = 0.0 !
! A31 A(12,2,14) 80.5627 -DE/DX = 0.0 !
! A32 A(13,2,14) 115.1638 -DE/DX = 0.0 !
! A33 A(4,3,11) 120.6613 -DE/DX = 0.0 !
! A34 A(4,3,12) 120.0232 -DE/DX = 0.0 !
! A35 A(4,3,13) 94.0558 -DE/DX = 0.0 !
! A36 A(4,3,14) 127.3507 -DE/DX = 0.0 !
! A37 A(11,3,12) 114.4864 -DE/DX = 0.0 !
! A38 A(11,3,13) 109.2312 -DE/DX = 0.0 !
! A39 A(11,3,14) 69.5135 -DE/DX = 0.0 !
! A40 A(12,3,13) 88.6205 -DE/DX = 0.0 !
! A41 A(12,3,14) 91.9441 -DE/DX = 0.0 !
! A42 A(13,3,14) 42.3516 -DE/DX = 0.0 !
! A43 A(2,4,5) 121.3465 -DE/DX = 0.0 !
! A44 A(2,4,6) 89.7903 -DE/DX = 0.0 !
! A45 A(3,4,5) 118.6679 -DE/DX = 0.0 !
! A46 A(3,4,6) 122.0341 -DE/DX = 0.0 !
! A47 A(5,4,6) 117.9096 -DE/DX = 0.0 !
! A48 A(1,6,4) 89.7901 -DE/DX = 0.0 !
! A49 A(1,6,8) 121.3467 -DE/DX = 0.0 !
! A50 A(4,6,7) 122.0341 -DE/DX = 0.0 !
! A51 A(4,6,8) 117.9096 -DE/DX = 0.0 !
! A52 A(7,6,8) 118.6679 -DE/DX = 0.0 !
! A53 A(6,7,9) 120.0232 -DE/DX = 0.0 !
! A54 A(6,7,10) 120.6613 -DE/DX = 0.0 !
! A55 A(6,7,15) 94.0558 -DE/DX = 0.0 !
! A56 A(6,7,16) 127.3506 -DE/DX = 0.0 !
! A57 A(9,7,10) 114.4864 -DE/DX = 0.0 !
! A58 A(9,7,15) 88.6207 -DE/DX = 0.0 !
! A59 A(9,7,16) 91.9443 -DE/DX = 0.0 !
! A60 A(10,7,15) 109.2309 -DE/DX = 0.0 !
! A61 A(10,7,16) 69.5133 -DE/DX = 0.0 !
! A62 A(15,7,16) 42.3515 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -20.742 -DE/DX = 0.0 !
! D2 D(6,1,2,4) 0.0 -DE/DX = 0.0 !
! D3 D(6,1,2,11) -45.918 -DE/DX = 0.0 !
! D4 D(6,1,2,12) -18.3309 -DE/DX = 0.0 !
! D5 D(6,1,2,13) 82.445 -DE/DX = 0.0 !
! D6 D(6,1,2,14) -123.2664 -DE/DX = 0.0 !
! D7 D(7,1,2,3) -0.0001 -DE/DX = 0.0 !
! D8 D(7,1,2,4) 20.742 -DE/DX = 0.0 !
! D9 D(7,1,2,11) -25.1761 -DE/DX = 0.0 !
! D10 D(7,1,2,12) 2.4111 -DE/DX = 0.0 !
! D11 D(7,1,2,13) 103.1869 -DE/DX = 0.0 !
! D12 D(7,1,2,14) -102.5244 -DE/DX = 0.0 !
! D13 D(9,1,2,3) -2.4112 -DE/DX = 0.0 !
! D14 D(9,1,2,4) 18.3308 -DE/DX = 0.0 !
! D15 D(9,1,2,11) -27.5872 -DE/DX = 0.0 !
! D16 D(9,1,2,12) -0.0001 -DE/DX = 0.0 !
! D17 D(9,1,2,13) 100.7758 -DE/DX = 0.0 !
! D18 D(9,1,2,14) -104.9356 -DE/DX = 0.0 !
! D19 D(10,1,2,3) 25.176 -DE/DX = 0.0 !
! D20 D(10,1,2,4) 45.918 -DE/DX = 0.0 !
! D21 D(10,1,2,11) -0.0001 -DE/DX = 0.0 !
! D22 D(10,1,2,12) 27.5871 -DE/DX = 0.0 !
! D23 D(10,1,2,13) 128.3629 -DE/DX = 0.0 !
! D24 D(10,1,2,14) -77.3484 -DE/DX = 0.0 !
! D25 D(15,1,2,3) -103.1869 -DE/DX = 0.0 !
! D26 D(15,1,2,4) -82.4449 -DE/DX = 0.0 !
! D27 D(15,1,2,11) -128.3629 -DE/DX = 0.0 !
! D28 D(15,1,2,12) -100.7758 -DE/DX = 0.0 !
! D29 D(15,1,2,13) 0.0 -DE/DX = 0.0 !
! D30 D(15,1,2,14) 154.2887 -DE/DX = 0.0 !
! D31 D(16,1,2,3) 102.5243 -DE/DX = 0.0 !
! D32 D(16,1,2,4) 123.2663 -DE/DX = 0.0 !
! D33 D(16,1,2,11) 77.3483 -DE/DX = 0.0 !
! D34 D(16,1,2,12) 104.9354 -DE/DX = 0.0 !
! D35 D(16,1,2,13) -154.2888 -DE/DX = 0.0 !
! D36 D(16,1,2,14) -0.0001 -DE/DX = 0.0 !
! D37 D(2,1,6,4) 0.0 -DE/DX = 0.0 !
! D38 D(2,1,6,8) -123.0828 -DE/DX = 0.0 !
! D39 D(9,1,6,4) -160.3592 -DE/DX = 0.0 !
! D40 D(9,1,6,8) 76.5581 -DE/DX = 0.0 !
! D41 D(10,1,6,4) -102.1158 -DE/DX = 0.0 !
! D42 D(10,1,6,8) 134.8014 -DE/DX = 0.0 !
! D43 D(15,1,6,4) 120.2482 -DE/DX = 0.0 !
! D44 D(15,1,6,8) -2.8345 -DE/DX = 0.0 !
! D45 D(16,1,6,4) -124.7021 -DE/DX = 0.0 !
! D46 D(16,1,6,8) 112.2151 -DE/DX = 0.0 !
! D47 D(1,2,4,5) 123.0828 -DE/DX = 0.0 !
! D48 D(1,2,4,6) 0.0 -DE/DX = 0.0 !
! D49 D(11,2,4,5) -134.8015 -DE/DX = 0.0 !
! D50 D(11,2,4,6) 102.1157 -DE/DX = 0.0 !
! D51 D(12,2,4,5) -76.5581 -DE/DX = 0.0 !
! D52 D(12,2,4,6) 160.3591 -DE/DX = 0.0 !
! D53 D(13,2,4,5) 2.8345 -DE/DX = 0.0 !
! D54 D(13,2,4,6) -120.2482 -DE/DX = 0.0 !
! D55 D(14,2,4,5) -112.2151 -DE/DX = 0.0 !
! D56 D(14,2,4,6) 124.7022 -DE/DX = 0.0 !
! D57 D(11,3,4,5) -160.592 -DE/DX = 0.0 !
! D58 D(11,3,4,6) 33.1448 -DE/DX = 0.0 !
! D59 D(12,3,4,5) -6.5509 -DE/DX = 0.0 !
! D60 D(12,3,4,6) -172.8141 -DE/DX = 0.0 !
! D61 D(13,3,4,5) 84.1997 -DE/DX = 0.0 !
! D62 D(13,3,4,6) -82.0635 -DE/DX = 0.0 !
! D63 D(14,3,4,5) 112.8115 -DE/DX = 0.0 !
! D64 D(14,3,4,6) -53.4517 -DE/DX = 0.0 !
! D65 D(2,4,6,1) 0.0 -DE/DX = 0.0 !
! D66 D(2,4,6,7) -40.4359 -DE/DX = 0.0 !
! D67 D(2,4,6,8) 125.9267 -DE/DX = 0.0 !
! D68 D(3,4,6,1) 40.436 -DE/DX = 0.0 !
! D69 D(3,4,6,7) 0.0 -DE/DX = 0.0 !
! D70 D(3,4,6,8) 166.3627 -DE/DX = 0.0 !
! D71 D(5,4,6,1) -125.9267 -DE/DX = 0.0 !
! D72 D(5,4,6,7) -166.3626 -DE/DX = 0.0 !
! D73 D(5,4,6,8) 0.0001 -DE/DX = 0.0 !
! D74 D(4,6,7,9) 172.8141 -DE/DX = 0.0 !
! D75 D(4,6,7,10) -33.1446 -DE/DX = 0.0 !
! D76 D(4,6,7,15) 82.0633 -DE/DX = 0.0 !
! D77 D(4,6,7,16) 53.4515 -DE/DX = 0.0 !
! D78 D(8,6,7,9) 6.5509 -DE/DX = 0.0 !
! D79 D(8,6,7,10) 160.5921 -DE/DX = 0.0 !
! D80 D(8,6,7,15) -84.2 -DE/DX = 0.0 !
! D81 D(8,6,7,16) -112.8118 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 524.81 cm-1. The corresponding vibrational mode is shown below, and the motion is similar to that of the HF/3-21G optimised structure but slower.
7. Thermochemistry
Sum of electronic and zero-point Energies= -234.403325 Sum of electronic and thermal Energies= -234.396907 Sum of electronic and thermal Enthalpies= -234.395963 Sum of electronic and thermal Free Energies= -234.432892
9. Key information
| Subject | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Transition state | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | B3LYP | 6-31G(d) | Default | -234.54389655 a.u. | Cs |
Diels Alder Reaction Between Cyclohexa-1,3-diene and Maleic Anhydride
Optimising the Reactants
(a) Optimisation of cyclohexa-1,3-diene
1. Optimising cyclohexa-1,3-diene using AM1 method
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000154 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.001022 0.001800 YES
RMS Displacement 0.000271 0.001200 YES
Predicted change in Energy=-1.882760D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.342 -DE/DX = 0.0 !
! R2 R(1,4) 1.4477 -DE/DX = 0.0002 !
! R3 R(1,5) 1.1 -DE/DX = 0.0 !
! R4 R(2,6) 1.1011 -DE/DX = 0.0 !
! R5 R(2,9) 1.4818 -DE/DX = -0.0001 !
! R6 R(3,4) 1.342 -DE/DX = 0.0 !
! R7 R(3,7) 1.1011 -DE/DX = 0.0 !
! R8 R(3,12) 1.4818 -DE/DX = -0.0001 !
! R9 R(4,8) 1.1 -DE/DX = 0.0 !
! R10 R(9,10) 1.1255 -DE/DX = 0.0 !
! R11 R(9,11) 1.1255 -DE/DX = 0.0 !
! R12 R(9,12) 1.5218 -DE/DX = -0.0001 !
! R13 R(12,13) 1.1255 -DE/DX = 0.0 !
! R14 R(12,14) 1.1255 -DE/DX = 0.0 !
! A1 A(2,1,4) 120.6853 -DE/DX = 0.0 !
! A2 A(2,1,5) 121.8282 -DE/DX = 0.0001 !
! A3 A(4,1,5) 117.4865 -DE/DX = 0.0 !
! A4 A(1,2,6) 121.351 -DE/DX = 0.0001 !
! A5 A(1,2,9) 123.3892 -DE/DX = 0.0 !
! A6 A(6,2,9) 115.2598 -DE/DX = 0.0 !
! A7 A(4,3,7) 121.3511 -DE/DX = 0.0001 !
! A8 A(4,3,12) 123.3892 -DE/DX = 0.0 !
! A9 A(7,3,12) 115.2598 -DE/DX = 0.0 !
! A10 A(1,4,3) 120.6853 -DE/DX = 0.0 !
! A11 A(1,4,8) 117.4865 -DE/DX = 0.0 !
! A12 A(3,4,8) 121.8282 -DE/DX = 0.0001 !
! A13 A(2,9,10) 108.072 -DE/DX = 0.0 !
! A14 A(2,9,11) 108.0596 -DE/DX = 0.0 !
! A15 A(2,9,12) 115.9255 -DE/DX = 0.0 !
! A16 A(10,9,11) 106.3594 -DE/DX = 0.0 !
! A17 A(10,9,12) 109.0064 -DE/DX = 0.0 !
! A18 A(11,9,12) 109.0031 -DE/DX = 0.0 !
! A19 A(3,12,9) 115.9255 -DE/DX = 0.0 !
! A20 A(3,12,13) 108.066 -DE/DX = 0.0 !
! A21 A(3,12,14) 108.0656 -DE/DX = 0.0 !
! A22 A(9,12,13) 109.0085 -DE/DX = 0.0 !
! A23 A(9,12,14) 109.0009 -DE/DX = 0.0 !
! A24 A(13,12,14) 106.3594 -DE/DX = 0.0 !
! D1 D(4,1,2,6) -179.9819 -DE/DX = 0.0 !
! D2 D(4,1,2,9) 0.0265 -DE/DX = 0.0 !
! D3 D(5,1,2,6) 0.01 -DE/DX = 0.0 !
! D4 D(5,1,2,9) -179.9816 -DE/DX = 0.0 !
! D5 D(2,1,4,3) 0.0186 -DE/DX = 0.0 !
! D6 D(2,1,4,8) -179.991 -DE/DX = 0.0 !
! D7 D(5,1,4,3) -179.9737 -DE/DX = 0.0 !
! D8 D(5,1,4,8) 0.0167 -DE/DX = 0.0 !
! D9 D(1,2,9,10) -122.7239 -DE/DX = 0.0 !
! D10 D(1,2,9,11) 122.5658 -DE/DX = 0.0 !
! D11 D(1,2,9,12) -0.072 -DE/DX = 0.0 !
! D12 D(6,2,9,10) 57.284 -DE/DX = 0.0 !
! D13 D(6,2,9,11) -57.4262 -DE/DX = 0.0 !
! D14 D(6,2,9,12) 179.9359 -DE/DX = 0.0 !
! D15 D(7,3,4,1) 179.9806 -DE/DX = 0.0 !
! D16 D(7,3,4,8) -0.0093 -DE/DX = 0.0 !
! D17 D(12,3,4,1) -0.0122 -DE/DX = 0.0 !
! D18 D(12,3,4,8) 179.9979 -DE/DX = 0.0 !
! D19 D(4,3,12,9) -0.035 -DE/DX = 0.0 !
! D20 D(4,3,12,13) 122.615 -DE/DX = 0.0 !
! D21 D(4,3,12,14) -122.6747 -DE/DX = 0.0 !
! D22 D(7,3,12,9) 179.9718 -DE/DX = 0.0 !
! D23 D(7,3,12,13) -57.3782 -DE/DX = 0.0 !
! D24 D(7,3,12,14) 57.3321 -DE/DX = 0.0 !
! D25 D(2,9,12,3) 0.0727 -DE/DX = 0.0 !
! D26 D(2,9,12,13) -122.0793 -DE/DX = 0.0 !
! D27 D(2,9,12,14) 122.2181 -DE/DX = 0.0 !
! D28 D(10,9,12,3) 122.2309 -DE/DX = 0.0 !
! D29 D(10,9,12,13) 0.0789 -DE/DX = 0.0 !
! D30 D(10,9,12,14) -115.6236 -DE/DX = 0.0 !
! D31 D(11,9,12,3) -122.0666 -DE/DX = 0.0 !
! D32 D(11,9,12,13) 115.7814 -DE/DX = 0.0 !
! D33 D(11,9,12,14) 0.0789 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Molecule | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Cyclohexa-1,3-diene | Optimisation to a minimum | Semi-empirical molecular orbital, AM1 | ZDO | Default | 0.02795812 a.u. | C2v |
(b) Optimisation of maleic anhydride
1. Optimising maleic anhydride using AM1 method
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000110 0.000450 YES
RMS Force 0.000027 0.000300 YES
Maximum Displacement 0.000207 0.001800 YES
RMS Displacement 0.000083 0.001200 YES
Predicted change in Energy=-3.291002D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4975 -DE/DX = 0.0 !
! R2 R(1,5) 1.4092 -DE/DX = 0.0 !
! R3 R(1,8) 1.2166 -DE/DX = -0.0001 !
! R4 R(2,3) 1.3487 -DE/DX = 0.0 !
! R5 R(2,6) 1.0905 -DE/DX = 0.0 !
! R6 R(3,4) 1.4975 -DE/DX = 0.0 !
! R7 R(3,7) 1.0905 -DE/DX = 0.0 !
! R8 R(4,5) 1.4092 -DE/DX = 0.0 !
! R9 R(4,9) 1.2165 -DE/DX = 0.0001 !
! A1 A(2,1,5) 108.2684 -DE/DX = 0.0 !
! A2 A(2,1,8) 134.6893 -DE/DX = 0.0 !
! A3 A(5,1,8) 117.0423 -DE/DX = 0.0 !
! A4 A(1,2,3) 107.9774 -DE/DX = 0.0 !
! A5 A(1,2,6) 121.6488 -DE/DX = 0.0 !
! A6 A(3,2,6) 130.3737 -DE/DX = 0.0 !
! A7 A(2,3,4) 107.9768 -DE/DX = 0.0 !
! A8 A(2,3,7) 130.3742 -DE/DX = 0.0 !
! A9 A(4,3,7) 121.6489 -DE/DX = 0.0 !
! A10 A(3,4,5) 108.2694 -DE/DX = 0.0 !
! A11 A(3,4,9) 134.6883 -DE/DX = 0.0 !
! A12 A(5,4,9) 117.0422 -DE/DX = 0.0 !
! A13 A(1,5,4) 107.508 -DE/DX = 0.0 !
! D1 D(5,1,2,3) -0.0039 -DE/DX = 0.0 !
! D2 D(5,1,2,6) -180.0016 -DE/DX = 0.0 !
! D3 D(8,1,2,3) 180.0008 -DE/DX = 0.0 !
! D4 D(8,1,2,6) 0.0031 -DE/DX = 0.0 !
! D5 D(2,1,5,4) 0.0013 -DE/DX = 0.0 !
! D6 D(8,1,5,4) -180.0024 -DE/DX = 0.0 !
! D7 D(1,2,3,4) 0.0047 -DE/DX = 0.0 !
! D8 D(1,2,3,7) 180.0023 -DE/DX = 0.0 !
! D9 D(6,2,3,4) 180.0021 -DE/DX = 0.0 !
! D10 D(6,2,3,7) -0.0003 -DE/DX = 0.0 !
! D11 D(2,3,4,5) -0.004 -DE/DX = 0.0 !
! D12 D(2,3,4,9) 180.0001 -DE/DX = 0.0 !
! D13 D(7,3,4,5) -180.0019 -DE/DX = 0.0 !
! D14 D(7,3,4,9) 0.0023 -DE/DX = 0.0 !
! D15 D(3,4,5,1) 0.0015 -DE/DX = 0.0 !
! D16 D(9,4,5,1) -180.0018 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
6. Key information
| Molecule | Job type | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|
| Maleic anhydride | Optimisation to a minimum | Semi-empirical molecular orbital, AM1 | ZDO | Default | -0.12182423 a.u. | C2v |
Optimising the Exo and Endo Transition Structures
(a) Optimisation of Exo transition state
1. Optimising exo transition state using AM1 method
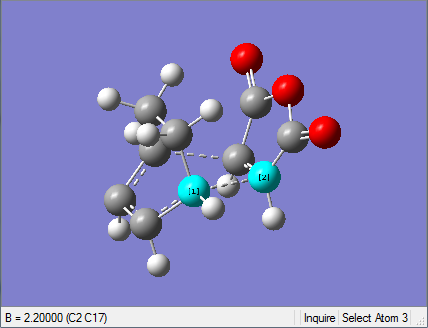
The guess exo transition state was drawn as above by combining the optimised cyclohexa-1,3-diene and maleic anhydride structures with two partially formed C-C bonds of 2.2 Å bond length and modifying the cyclohexa-1,3-diene into envelope structre. The optimised structure is shown below, which has 2.17078 Å bond lengths for the partially formed bonds.
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000120 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.001573 0.001800 YES
RMS Displacement 0.000381 0.001200 YES
Predicted change in Energy=-8.526949D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3943 -DE/DX = 0.0 !
! R2 R(1,4) 1.3967 -DE/DX = 0.0001 !
! R3 R(1,5) 1.0995 -DE/DX = 0.0 !
! R4 R(2,8) 1.1022 -DE/DX = 0.0 !
! R5 R(2,12) 1.4898 -DE/DX = 0.0 !
! R6 R(2,17) 2.1708 -DE/DX = 0.0 !
! R7 R(3,4) 1.3944 -DE/DX = 0.0 !
! R8 R(3,7) 1.1022 -DE/DX = 0.0 !
! R9 R(3,9) 1.4898 -DE/DX = 0.0 !
! R10 R(3,16) 2.17 -DE/DX = 0.0 !
! R11 R(4,6) 1.0995 -DE/DX = 0.0 !
! R12 R(9,10) 1.124 -DE/DX = 0.0 !
! R13 R(9,11) 1.1262 -DE/DX = 0.0 !
! R14 R(9,12) 1.5221 -DE/DX = 0.0 !
! R15 R(12,13) 1.124 -DE/DX = 0.0 !
! R16 R(12,14) 1.1262 -DE/DX = 0.0 !
! R17 R(15,16) 1.4882 -DE/DX = 0.0 !
! R18 R(15,19) 1.4096 -DE/DX = 0.0 !
! R19 R(15,22) 1.2206 -DE/DX = 0.0 !
! R20 R(16,17) 1.41 -DE/DX = 0.0001 !
! R21 R(16,20) 1.0926 -DE/DX = 0.0 !
! R22 R(17,18) 1.4882 -DE/DX = 0.0 !
! R23 R(17,21) 1.0926 -DE/DX = 0.0 !
! R24 R(18,19) 1.4097 -DE/DX = 0.0 !
! R25 R(18,23) 1.2206 -DE/DX = -0.0001 !
! A1 A(2,1,4) 118.1251 -DE/DX = 0.0 !
! A2 A(2,1,5) 120.7728 -DE/DX = 0.0 !
! A3 A(4,1,5) 120.3832 -DE/DX = 0.0 !
! A4 A(1,2,8) 120.4887 -DE/DX = 0.0 !
! A5 A(1,2,12) 119.6892 -DE/DX = 0.0 !
! A6 A(1,2,17) 92.7345 -DE/DX = 0.0 !
! A7 A(8,2,12) 115.8574 -DE/DX = 0.0 !
! A8 A(8,2,17) 97.5619 -DE/DX = 0.0 !
! A9 A(12,2,17) 99.7893 -DE/DX = 0.0 !
! A10 A(4,3,7) 120.4896 -DE/DX = 0.0 !
! A11 A(4,3,9) 119.6761 -DE/DX = 0.0 !
! A12 A(4,3,16) 92.7503 -DE/DX = 0.0 !
! A13 A(7,3,9) 115.8531 -DE/DX = 0.0 !
! A14 A(7,3,16) 97.5506 -DE/DX = 0.0 !
! A15 A(9,3,16) 99.8269 -DE/DX = 0.0 !
! A16 A(1,4,3) 118.1104 -DE/DX = 0.0 !
! A17 A(1,4,6) 120.3904 -DE/DX = 0.0 !
! A18 A(3,4,6) 120.7776 -DE/DX = 0.0 !
! A19 A(3,9,10) 110.2442 -DE/DX = 0.0 !
! A20 A(3,9,11) 107.3115 -DE/DX = 0.0 !
! A21 A(3,9,12) 113.5186 -DE/DX = 0.0 !
! A22 A(10,9,11) 106.2917 -DE/DX = 0.0 !
! A23 A(10,9,12) 110.0253 -DE/DX = 0.0 !
! A24 A(11,9,12) 109.1547 -DE/DX = 0.0 !
! A25 A(2,12,9) 113.5172 -DE/DX = 0.0 !
! A26 A(2,12,13) 110.2472 -DE/DX = 0.0 !
! A27 A(2,12,14) 107.3179 -DE/DX = 0.0 !
! A28 A(9,12,13) 110.0228 -DE/DX = 0.0 !
! A29 A(9,12,14) 109.1596 -DE/DX = 0.0 !
! A30 A(13,12,14) 106.2809 -DE/DX = 0.0 !
! A31 A(16,15,19) 109.0509 -DE/DX = 0.0 !
! A32 A(16,15,22) 134.8473 -DE/DX = 0.0 !
! A33 A(19,15,22) 116.1017 -DE/DX = 0.0 !
! A34 A(3,16,15) 99.5856 -DE/DX = 0.0 !
! A35 A(3,16,17) 107.447 -DE/DX = 0.0 !
! A36 A(3,16,20) 89.6269 -DE/DX = 0.0 !
! A37 A(15,16,17) 106.9854 -DE/DX = 0.0 !
! A38 A(15,16,20) 120.414 -DE/DX = 0.0 !
! A39 A(17,16,20) 125.9738 -DE/DX = 0.0 !
! A40 A(2,17,16) 107.4327 -DE/DX = 0.0 !
! A41 A(2,17,18) 99.5939 -DE/DX = 0.0 !
! A42 A(2,17,21) 89.6056 -DE/DX = 0.0 !
! A43 A(16,17,18) 106.9892 -DE/DX = 0.0 !
! A44 A(16,17,21) 125.9873 -DE/DX = 0.0 !
! A45 A(18,17,21) 120.4114 -DE/DX = 0.0 !
! A46 A(17,18,19) 109.049 -DE/DX = 0.0 !
! A47 A(17,18,23) 134.8508 -DE/DX = 0.0 !
! A48 A(19,18,23) 116.1 -DE/DX = 0.0 !
! A49 A(15,19,18) 107.9169 -DE/DX = 0.0 !
! D1 D(4,1,2,8) -168.9629 -DE/DX = 0.0 !
! D2 D(4,1,2,12) 34.3611 -DE/DX = 0.0 !
! D3 D(4,1,2,17) -68.5343 -DE/DX = 0.0 !
! D4 D(5,1,2,8) 1.3593 -DE/DX = 0.0 !
! D5 D(5,1,2,12) -155.3167 -DE/DX = 0.0 !
! D6 D(5,1,2,17) 101.7878 -DE/DX = 0.0 !
! D7 D(2,1,4,3) 0.001 -DE/DX = 0.0 !
! D8 D(2,1,4,6) 170.3452 -DE/DX = 0.0 !
! D9 D(5,1,4,3) -170.3604 -DE/DX = 0.0 !
! D10 D(5,1,4,6) -0.0161 -DE/DX = 0.0 !
! D11 D(1,2,12,9) -32.8475 -DE/DX = 0.0 !
! D12 D(1,2,12,13) -156.8011 -DE/DX = 0.0 !
! D13 D(1,2,12,14) 87.8573 -DE/DX = 0.0 !
! D14 D(8,2,12,9) 169.4331 -DE/DX = 0.0 !
! D15 D(8,2,12,13) 45.4795 -DE/DX = 0.0 !
! D16 D(8,2,12,14) -69.8621 -DE/DX = 0.0 !
! D17 D(17,2,12,9) 66.0171 -DE/DX = 0.0 !
! D18 D(17,2,12,13) -57.9365 -DE/DX = 0.0 !
! D19 D(17,2,12,14) -173.2782 -DE/DX = 0.0 !
! D20 D(1,2,17,16) 59.3625 -DE/DX = 0.0 !
! D21 D(1,2,17,18) 170.6887 -DE/DX = 0.0 !
! D22 D(1,2,17,21) -68.4985 -DE/DX = 0.0 !
! D23 D(8,2,17,16) -179.3895 -DE/DX = 0.0 !
! D24 D(8,2,17,18) -68.0632 -DE/DX = 0.0 !
! D25 D(8,2,17,21) 52.7496 -DE/DX = 0.0 !
! D26 D(12,2,17,16) -61.3965 -DE/DX = 0.0 !
! D27 D(12,2,17,18) 49.9298 -DE/DX = 0.0 !
! D28 D(12,2,17,21) 170.7426 -DE/DX = 0.0 !
! D29 D(7,3,4,1) 168.978 -DE/DX = 0.0 !
! D30 D(7,3,4,6) -1.3272 -DE/DX = 0.0 !
! D31 D(9,3,4,1) -34.3934 -DE/DX = 0.0 !
! D32 D(9,3,4,6) 155.3014 -DE/DX = 0.0 !
! D33 D(16,3,4,1) 68.5529 -DE/DX = 0.0 !
! D34 D(16,3,4,6) -101.7523 -DE/DX = 0.0 !
! D35 D(4,3,9,10) 156.8999 -DE/DX = 0.0 !
! D36 D(4,3,9,11) -87.7508 -DE/DX = 0.0 !
! D37 D(4,3,9,12) 32.9443 -DE/DX = 0.0 !
! D38 D(7,3,9,10) -45.4247 -DE/DX = 0.0 !
! D39 D(7,3,9,11) 69.9246 -DE/DX = 0.0 !
! D40 D(7,3,9,12) -169.3803 -DE/DX = 0.0 !
! D41 D(16,3,9,10) 57.9975 -DE/DX = 0.0 !
! D42 D(16,3,9,11) 173.3467 -DE/DX = 0.0 !
! D43 D(16,3,9,12) -65.9581 -DE/DX = 0.0 !
! D44 D(4,3,16,15) -170.6853 -DE/DX = 0.0 !
! D45 D(4,3,16,17) -59.3622 -DE/DX = 0.0 !
! D46 D(4,3,16,20) 68.4959 -DE/DX = 0.0 !
! D47 D(7,3,16,15) 68.0644 -DE/DX = 0.0 !
! D48 D(7,3,16,17) 179.3875 -DE/DX = 0.0 !
! D49 D(7,3,16,20) -52.7544 -DE/DX = 0.0 !
! D50 D(9,3,16,15) -49.9303 -DE/DX = 0.0 !
! D51 D(9,3,16,17) 61.3928 -DE/DX = 0.0 !
! D52 D(9,3,16,20) -170.749 -DE/DX = 0.0 !
! D53 D(3,9,12,2) -0.0594 -DE/DX = 0.0 !
! D54 D(3,9,12,13) 124.0161 -DE/DX = 0.0 !
! D55 D(3,9,12,14) -119.7207 -DE/DX = 0.0 !
! D56 D(10,9,12,2) -124.1339 -DE/DX = 0.0 !
! D57 D(10,9,12,13) -0.0584 -DE/DX = 0.0 !
! D58 D(10,9,12,14) 116.2048 -DE/DX = 0.0 !
! D59 D(11,9,12,2) 119.5912 -DE/DX = 0.0 !
! D60 D(11,9,12,13) -116.3333 -DE/DX = 0.0 !
! D61 D(11,9,12,14) -0.07 -DE/DX = 0.0 !
! D62 D(19,15,16,3) 111.1286 -DE/DX = 0.0 !
! D63 D(19,15,16,17) -0.5575 -DE/DX = 0.0 !
! D64 D(19,15,16,20) -153.6227 -DE/DX = 0.0 !
! D65 D(22,15,16,3) -69.0334 -DE/DX = 0.0 !
! D66 D(22,15,16,17) 179.2805 -DE/DX = 0.0 !
! D67 D(22,15,16,20) 26.2153 -DE/DX = 0.0 !
! D68 D(16,15,19,18) 0.915 -DE/DX = 0.0 !
! D69 D(22,15,19,18) -178.9571 -DE/DX = 0.0 !
! D70 D(3,16,17,2) -0.0033 -DE/DX = 0.0 !
! D71 D(3,16,17,18) -106.1796 -DE/DX = 0.0 !
! D72 D(3,16,17,21) 102.6539 -DE/DX = 0.0 !
! D73 D(15,16,17,2) 106.1681 -DE/DX = 0.0 !
! D74 D(15,16,17,18) -0.0083 -DE/DX = 0.0 !
! D75 D(15,16,17,21) -151.1748 -DE/DX = 0.0 !
! D76 D(20,16,17,2) -102.6938 -DE/DX = 0.0 !
! D77 D(20,16,17,18) 151.1298 -DE/DX = 0.0 !
! D78 D(20,16,17,21) -0.0366 -DE/DX = 0.0 !
! D79 D(2,17,18,19) -111.1034 -DE/DX = 0.0 !
! D80 D(2,17,18,23) 69.0593 -DE/DX = 0.0 !
! D81 D(16,17,18,19) 0.5715 -DE/DX = 0.0 !
! D82 D(16,17,18,23) -179.2657 -DE/DX = 0.0 !
! D83 D(21,17,18,19) 153.6686 -DE/DX = 0.0 !
! D84 D(21,17,18,23) -26.1687 -DE/DX = 0.0 !
! D85 D(17,18,19,15) -0.9202 -DE/DX = 0.0 !
! D86 D(23,18,19,15) 178.9513 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 812.23 cm-1. The corresponding vibrational mode is shown below. Although the reactants different, the transition structure for reaction between cyclohexa-1,3-diene and maleic Anhydride has a similar vibrational motion as the transition state structure for reaction between cis-Butadiene and ethylene obtained earlier, that is, the two C atoms of maleic Anhydride and the two middle C atoms of cyclohexa-1,3-diene approach each other in a sychronised motion and facilitates two simultaneous C-C bonds formation.
7. Thermochemistry
Sum of electronic and zero-point Energies= 0.134881 Sum of electronic and thermal Energies= 0.144882 Sum of electronic and thermal Enthalpies= 0.145826 Sum of electronic and thermal Free Energies= 0.099117
8. Key information
| Transition state type | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Exo | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | Semi-empirical molecular orbital, AM1 | ZDO | Default | -0.05041976 a.u. | Cs |
(b) Optimisation of Endo transition state
1. Optimising endo transition state using AM1 method
The guess endo transition state was drawn in a similar way as for exo transition state.
test molecule |
2. File link
The optimised file is linked to here.
The gradient is less than 0.001, which means the optimisation is complete.
4. Real output
Item Value Threshold Converged?
Maximum Force 0.000013 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.001115 0.001800 YES
RMS Displacement 0.000199 0.001200 YES
Predicted change in Energy=-1.746014D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.393 -DE/DX = 0.0 !
! R2 R(1,4) 1.3972 -DE/DX = 0.0 !
! R3 R(1,5) 1.1006 -DE/DX = 0.0 !
! R4 R(2,8) 1.1024 -DE/DX = 0.0 !
! R5 R(2,12) 1.4905 -DE/DX = 0.0 !
! R6 R(2,16) 2.1623 -DE/DX = 0.0 !
! R7 R(3,4) 1.3931 -DE/DX = 0.0 !
! R8 R(3,7) 1.1024 -DE/DX = 0.0 !
! R9 R(3,9) 1.4905 -DE/DX = 0.0 !
! R10 R(3,17) 2.1624 -DE/DX = 0.0 !
! R11 R(4,6) 1.1006 -DE/DX = 0.0 !
! R12 R(9,10) 1.1224 -DE/DX = 0.0 !
! R13 R(9,11) 1.1261 -DE/DX = 0.0 !
! R14 R(9,12) 1.523 -DE/DX = 0.0 !
! R15 R(12,13) 1.1224 -DE/DX = 0.0 !
! R16 R(12,14) 1.1261 -DE/DX = 0.0 !
! R17 R(15,16) 1.4892 -DE/DX = 0.0 !
! R18 R(15,19) 1.409 -DE/DX = 0.0 !
! R19 R(15,22) 1.2206 -DE/DX = 0.0 !
! R20 R(16,17) 1.4085 -DE/DX = 0.0 !
! R21 R(16,20) 1.0929 -DE/DX = 0.0 !
! R22 R(17,18) 1.4892 -DE/DX = 0.0 !
! R23 R(17,21) 1.0929 -DE/DX = 0.0 !
! R24 R(18,19) 1.409 -DE/DX = 0.0 !
! R25 R(18,23) 1.2206 -DE/DX = 0.0 !
! A1 A(2,1,4) 118.2165 -DE/DX = 0.0 !
! A2 A(2,1,5) 120.7313 -DE/DX = 0.0 !
! A3 A(4,1,5) 120.3283 -DE/DX = 0.0 !
! A4 A(1,2,8) 119.9715 -DE/DX = 0.0 !
! A5 A(1,2,12) 119.926 -DE/DX = 0.0 !
! A6 A(1,2,16) 96.7578 -DE/DX = 0.0 !
! A7 A(8,2,12) 116.2572 -DE/DX = 0.0 !
! A8 A(8,2,16) 98.0361 -DE/DX = 0.0 !
! A9 A(12,2,16) 94.8221 -DE/DX = 0.0 !
! A10 A(4,3,7) 119.972 -DE/DX = 0.0 !
! A11 A(4,3,9) 119.9187 -DE/DX = 0.0 !
! A12 A(4,3,17) 96.754 -DE/DX = 0.0 !
! A13 A(7,3,9) 116.259 -DE/DX = 0.0 !
! A14 A(7,3,17) 98.0409 -DE/DX = 0.0 !
! A15 A(9,3,17) 94.8341 -DE/DX = 0.0 !
! A16 A(1,4,3) 118.2165 -DE/DX = 0.0 !
! A17 A(1,4,6) 120.3281 -DE/DX = 0.0 !
! A18 A(3,4,6) 120.7314 -DE/DX = 0.0 !
! A19 A(3,9,10) 110.0842 -DE/DX = 0.0 !
! A20 A(3,9,11) 107.4557 -DE/DX = 0.0 !
! A21 A(3,9,12) 113.5597 -DE/DX = 0.0 !
! A22 A(10,9,11) 106.4376 -DE/DX = 0.0 !
! A23 A(10,9,12) 109.9444 -DE/DX = 0.0 !
! A24 A(11,9,12) 109.0785 -DE/DX = 0.0 !
! A25 A(2,12,9) 113.5599 -DE/DX = 0.0 !
! A26 A(2,12,13) 110.0786 -DE/DX = 0.0 !
! A27 A(2,12,14) 107.4573 -DE/DX = 0.0 !
! A28 A(9,12,13) 109.9455 -DE/DX = 0.0 !
! A29 A(9,12,14) 109.0767 -DE/DX = 0.0 !
! A30 A(13,12,14) 106.4425 -DE/DX = 0.0 !
! A31 A(16,15,19) 109.018 -DE/DX = 0.0 !
! A32 A(16,15,22) 134.7615 -DE/DX = 0.0 !
! A33 A(19,15,22) 116.2183 -DE/DX = 0.0 !
! A34 A(2,16,15) 100.0256 -DE/DX = 0.0 !
! A35 A(2,16,17) 107.5789 -DE/DX = 0.0 !
! A36 A(2,16,20) 88.616 -DE/DX = 0.0 !
! A37 A(15,16,17) 106.9979 -DE/DX = 0.0 !
! A38 A(15,16,20) 120.5094 -DE/DX = 0.0 !
! A39 A(17,16,20) 126.1489 -DE/DX = 0.0 !
! A40 A(3,17,16) 107.5757 -DE/DX = 0.0 !
! A41 A(3,17,18) 100.0246 -DE/DX = 0.0 !
! A42 A(3,17,21) 88.624 -DE/DX = 0.0 !
! A43 A(16,17,18) 106.9995 -DE/DX = 0.0 !
! A44 A(16,17,21) 126.1471 -DE/DX = 0.0 !
! A45 A(18,17,21) 120.5072 -DE/DX = 0.0 !
! A46 A(17,18,19) 109.0173 -DE/DX = 0.0 !
! A47 A(17,18,23) 134.7619 -DE/DX = 0.0 !
! A48 A(19,18,23) 116.2186 -DE/DX = 0.0 !
! A49 A(15,19,18) 107.9644 -DE/DX = 0.0 !
! D1 D(4,1,2,8) -169.2253 -DE/DX = 0.0 !
! D2 D(4,1,2,12) 33.6698 -DE/DX = 0.0 !
! D3 D(4,1,2,16) -65.8787 -DE/DX = 0.0 !
! D4 D(5,1,2,8) 1.0586 -DE/DX = 0.0 !
! D5 D(5,1,2,12) -156.0462 -DE/DX = 0.0 !
! D6 D(5,1,2,16) 104.4053 -DE/DX = 0.0 !
! D7 D(2,1,4,3) -0.0016 -DE/DX = 0.0 !
! D8 D(2,1,4,6) 170.3222 -DE/DX = 0.0 !
! D9 D(5,1,4,3) -170.3262 -DE/DX = 0.0 !
! D10 D(5,1,4,6) -0.0024 -DE/DX = 0.0 !
! D11 D(1,2,12,9) -32.1894 -DE/DX = 0.0 !
! D12 D(1,2,12,13) -155.9427 -DE/DX = 0.0 !
! D13 D(1,2,12,14) 88.5345 -DE/DX = 0.0 !
! D14 D(8,2,12,9) 169.8841 -DE/DX = 0.0 !
! D15 D(8,2,12,13) 46.1309 -DE/DX = 0.0 !
! D16 D(8,2,12,14) -69.392 -DE/DX = 0.0 !
! D17 D(16,2,12,9) 68.4611 -DE/DX = 0.0 !
! D18 D(16,2,12,13) -55.2922 -DE/DX = 0.0 !
! D19 D(16,2,12,14) -170.815 -DE/DX = 0.0 !
! D20 D(1,2,16,15) -54.0347 -DE/DX = 0.0 !
! D21 D(1,2,16,17) 57.522 -DE/DX = 0.0 !
! D22 D(1,2,16,20) -174.7939 -DE/DX = 0.0 !
! D23 D(8,2,16,15) 67.6189 -DE/DX = 0.0 !
! D24 D(8,2,16,17) 179.1756 -DE/DX = 0.0 !
! D25 D(8,2,16,20) -53.1403 -DE/DX = 0.0 !
! D26 D(12,2,16,15) -174.9754 -DE/DX = 0.0 !
! D27 D(12,2,16,17) -63.4188 -DE/DX = 0.0 !
! D28 D(12,2,16,20) 64.2653 -DE/DX = 0.0 !
! D29 D(7,3,4,1) 169.228 -DE/DX = 0.0 !
! D30 D(7,3,4,6) -1.0552 -DE/DX = 0.0 !
! D31 D(9,3,4,1) -33.6809 -DE/DX = 0.0 !
! D32 D(9,3,4,6) 156.0359 -DE/DX = 0.0 !
! D33 D(17,3,4,1) 65.8778 -DE/DX = 0.0 !
! D34 D(17,3,4,6) -104.4054 -DE/DX = 0.0 !
! D35 D(4,3,9,10) 155.987 -DE/DX = 0.0 !
! D36 D(4,3,9,11) -88.494 -DE/DX = 0.0 !
! D37 D(4,3,9,12) 32.231 -DE/DX = 0.0 !
! D38 D(7,3,9,10) -46.1 -DE/DX = 0.0 !
! D39 D(7,3,9,11) 69.4191 -DE/DX = 0.0 !
! D40 D(7,3,9,12) -169.8559 -DE/DX = 0.0 !
! D41 D(17,3,9,10) 55.335 -DE/DX = 0.0 !
! D42 D(17,3,9,11) 170.8541 -DE/DX = 0.0 !
! D43 D(17,3,9,12) -68.4209 -DE/DX = 0.0 !
! D44 D(4,3,17,16) -57.5248 -DE/DX = 0.0 !
! D45 D(4,3,17,18) 54.0322 -DE/DX = 0.0 !
! D46 D(4,3,17,21) 174.7907 -DE/DX = 0.0 !
! D47 D(7,3,17,16) -179.1791 -DE/DX = 0.0 !
! D48 D(7,3,17,18) -67.6221 -DE/DX = 0.0 !
! D49 D(7,3,17,21) 53.1364 -DE/DX = 0.0 !
! D50 D(9,3,17,16) 63.4102 -DE/DX = 0.0 !
! D51 D(9,3,17,18) 174.9673 -DE/DX = 0.0 !
! D52 D(9,3,17,21) -64.2743 -DE/DX = 0.0 !
! D53 D(3,9,12,2) -0.0271 -DE/DX = 0.0 !
! D54 D(3,9,12,13) 123.7987 -DE/DX = 0.0 !
! D55 D(3,9,12,14) -119.8349 -DE/DX = 0.0 !
! D56 D(10,9,12,2) -123.8591 -DE/DX = 0.0 !
! D57 D(10,9,12,13) -0.0334 -DE/DX = 0.0 !
! D58 D(10,9,12,14) 116.333 -DE/DX = 0.0 !
! D59 D(11,9,12,2) 119.78 -DE/DX = 0.0 !
! D60 D(11,9,12,13) -116.3943 -DE/DX = 0.0 !
! D61 D(11,9,12,14) -0.0279 -DE/DX = 0.0 !
! D62 D(19,15,16,2) 111.683 -DE/DX = 0.0 !
! D63 D(19,15,16,17) -0.3262 -DE/DX = 0.0 !
! D64 D(19,15,16,20) -153.9752 -DE/DX = 0.0 !
! D65 D(22,15,16,2) -68.8997 -DE/DX = 0.0 !
! D66 D(22,15,16,17) 179.0912 -DE/DX = 0.0 !
! D67 D(22,15,16,20) 25.4421 -DE/DX = 0.0 !
! D68 D(16,15,19,18) 0.5266 -DE/DX = 0.0 !
! D69 D(22,15,19,18) -179.0122 -DE/DX = 0.0 !
! D70 D(2,16,17,3) 0.0018 -DE/DX = 0.0 !
! D71 D(2,16,17,18) -106.723 -DE/DX = 0.0 !
! D72 D(2,16,17,21) 101.5494 -DE/DX = 0.0 !
! D73 D(15,16,17,3) 106.7284 -DE/DX = 0.0 !
! D74 D(15,16,17,18) 0.0036 -DE/DX = 0.0 !
! D75 D(15,16,17,21) -151.7239 -DE/DX = 0.0 !
! D76 D(20,16,17,3) -101.5388 -DE/DX = 0.0 !
! D77 D(20,16,17,18) 151.7364 -DE/DX = 0.0 !
! D78 D(20,16,17,21) 0.0089 -DE/DX = 0.0 !
! D79 D(3,17,18,19) -111.6856 -DE/DX = 0.0 !
! D80 D(3,17,18,23) 68.8971 -DE/DX = 0.0 !
! D81 D(16,17,18,19) 0.3201 -DE/DX = 0.0 !
! D82 D(16,17,18,23) -179.0971 -DE/DX = 0.0 !
! D83 D(21,17,18,19) 153.9643 -DE/DX = 0.0 !
! D84 D(21,17,18,23) -25.453 -DE/DX = 0.0 !
! D85 D(17,18,19,15) -0.5244 -DE/DX = 0.0 !
! D86 D(23,18,19,15) 179.0144 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Both force and displacement are converged, indicating the success of optimisation.
As the above table is shown, there is only one imaginary frequency that has a magnitude of 806.40 cm-1. The corresponding vibrational mode is shown below, and the endo transition structure has a similar vibrational motion as the exo transition structure obtained earlier.
7. Thermochemistry
Sum of electronic and zero-point Energies= 0.133495 Sum of electronic and thermal Energies= 0.143684 Sum of electronic and thermal Enthalpies= 0.144628 Sum of electronic and thermal Free Energies= 0.097351
8. Key information
| Transition state type | Job type | Additional keywords | Method | Basis set | Memory limit | Energy | Point group |
|---|---|---|---|---|---|---|---|
| Endo | Optimisation to a TS (Berny), calculate the force constants once | Opt=NoEigen | Semi-empirical molecular orbital, AM1 | ZDO | Default | -0.05150479 a.u. | Cs |
(c) HOMO visialisation
They are both antisymmterical.