Rep:Mod:yudanchen
Week 1 Compulsory Section
Optimisation Analysis
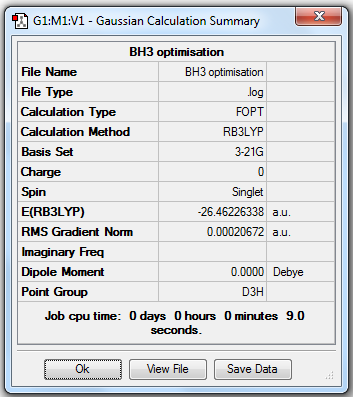
BH3 Optimisation (3-21G)
1. Optimising BH3 Using Basis Set 3-21G
Before optimisation, BH3 was set as: B-H bond length = 1.5Å, H-B-H bond angle = 120o.
Then the molecule was optimised using B3LYP as the calculation method and 3-21G as the basis set.

2. File Link
The file of the optimisation is here.
3. Geometric Information
B-H bond length = 1.19349Å, H-B-H bond angle = 120o.
5. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
6. Optimisation Plot

The following Total Energy curve shows how the energy of BH3 molecule changes through optimisation. The Gaussview Program traversed the potential energy surface of BH3 and found the minimum energy structure.
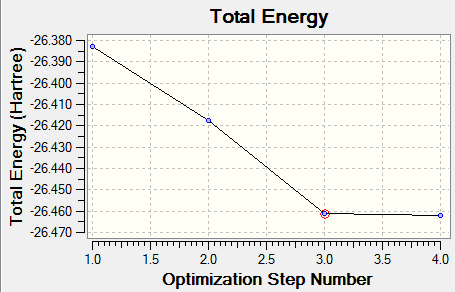
The following Root Mean Square Gradient curve gives the gradient of the energy of BH3 molecule at each optimisation step. It shows the gradient going to zero as we approach the minimum energy.
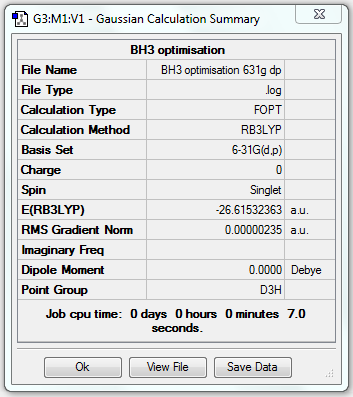
BH3 Optimisation (6-31G(d,p))
1. Optimising BH3 Using Basis Set 6-31G(d,p)

2. File Link
The file of the optimisation is here.
3. Geometric Information
B-H bond length = 1.19232Å, H-B-H bond angle = 120o.
5. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
6. Reporting Total Energies
| BH3(3-21G) | BH3(6-31G(d,p)) | |
|---|---|---|
| Total energy | -26.46226338a.u. | -26.61532363a.u. |
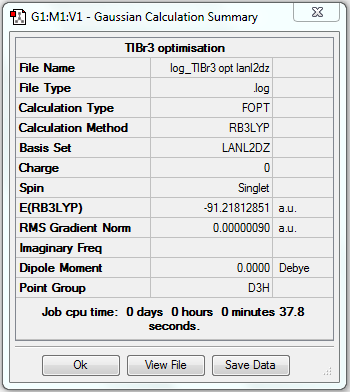
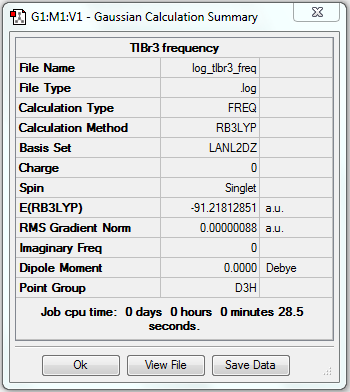
TlBr3 Optimisation (LanL2DZ)
1. Optimising TlBr3 Using Pseudo-potentials and Larger Basis Set
2. D-space Link
The D-space link of the Optimisation is [1].
3. Geometric Information
Tl-Br bond length = 2.65095Å, Br-Tl-Br bond angle = 120o.
5. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084121D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
6. Comparison to Literature
| LanL2DZ optimised | Literature | |
|---|---|---|
| Tl-Br bond distance | 2.65095Å | 2.564Å[1] |
The literature value is only slightly smaller than the experimental value after optimisation, which shows the result is reasonable.
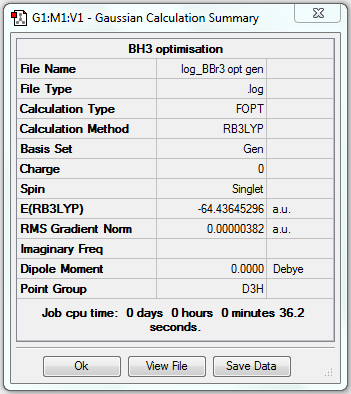
BBr3 Optimisation (Mixed PPs & Basis Sets)
1. Optimising BBr3 Using A Mixture of Pseudo-potentials and Basis Sets
2. D-space Link
The D-space link of the Optimisation is [2].
3. Geometric Information
B-Br bond length = 1.93396Å, Br-B-Br bond angle = 120o.
5. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027599D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Result Analysis
1. Bond Distance Comparison
| Molecule | Bond distance |
|---|---|
| BH3 (3-21G) | 1.19349Å |
| BH3 (6-31G(d,p)) | 1.19232Å |
| BBr3 | 1.93396Å |
| TlBr3 | 2.65095Å |
Bond distance is the length between the centers of two atoms within a molecule, and it depends on size, electronegativity, and degree of overlap of the two atoms. As the ligands change from H to Br, the bond distance increases, which is mainly because Br is much larger than H and the overlap is weaker for BBr than BH, and thus the bond strength is weaker for B-Br than B-H. H and Br have similar electronegativity though, and the slightly larger electronegativity difference between B and Br weakens the B-Br covalent bond further. When the central element is changed from B to Tl, the bond distance increases. Although B and Tl have exactly the same electronegativity, Tl is much larger than B and overlap of TlBr is weaker than BBr, therefore Tl-Br bond is weaker than B-Br bond.
2. Questions
(1) In some structures GaussView does not draw in the bonds where we expect, does this mean there is no bond? Why?
Answer: No. GaussView draws bonds based on a distance standard. If the bond distance exceeds that pre-defined value, the bonds will not show but they still exist.
(2) What is a bond?
Answer: A bond is the attraction holding atoms together.
Frequency Analysis
Frequency Analysis of BH3
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
4. Vibrational Analysis
5. IR Spectrum

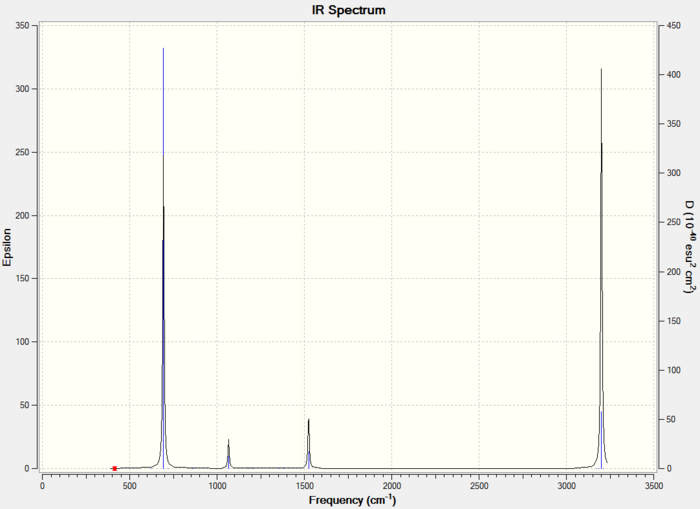
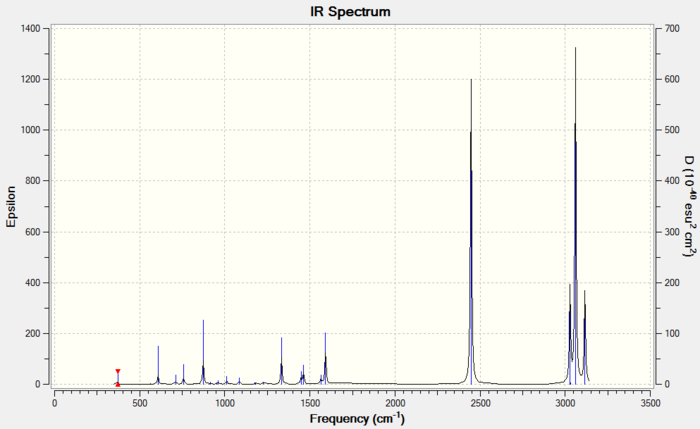
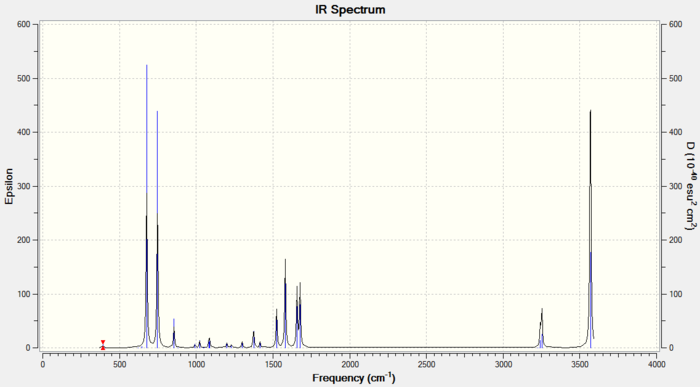
The fourth vibration has zero intensity leading to a missing peak, the second and third vibrations have the same frequency leading to only one peak, and similarly the fifth and sixth vibrations giving only one peak. Plus the first vibration gives one peak, there are 3 peaks in total, which are shown in the IR spectrum.
Frequency Analysis of TlBr3
1. D-space Link
The D-space link of the frequency analysis is [3].
3. "Real" Output
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
4. Vibrational Frequency Comparison
| No. | Vibrational frequency of BH3 | Vibrational frequency of TlBr3 | Symmetry of BH3 | Symmtery of TlBr3 |
|---|---|---|---|---|
| 1 | 1163.00 | 46.43 | A2' | E' |
| 2 | 1213.19 | 46.43 | E' | E' |
| 3 | 1213.19 | 52.14 | E' | A2' |
| 4 | 2582.26 | 165.27 | A1' | A1' |
| 5 | 2715.43 | 210.69 | E' | E' |
| 6 | 2715.43 | 210.69 | E' | E' |
We know that v=(k/m)0.5 where v=vibrational frequency, k=force constant and m=reduced mass. There are two reasons contributing to the large difference in vibrational frequency between BH3 and TlBr3. Firstly, as it is mentioned previously, Tl-Br bond is much weaker than B-H bond, leading to smaller k and thus lower vibrational frequency for TlBr3. Secondly, Tl and Br are heavier atoms than B and H, therefore m is larger and thus again lower vibrational frequency for TlBr3.
There has been a reordering of modes. For BH3 the lowest frequency mode is A2', while the lowest frequency mode is the degenerate E' for TlBr3.
5. IR Spectrum
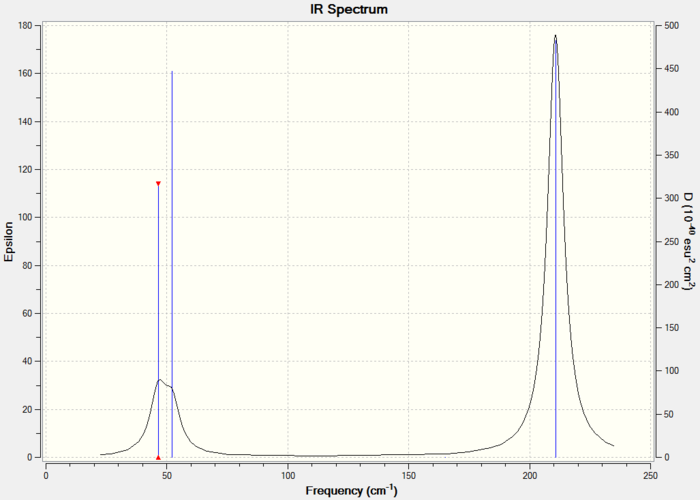
This IR spectrum has 3 peaks just like the one for BH3.
6. Questions
(1) Why must you use the same method and basis set for both the optimisation and frequency analysis calculations?
Answer: The accuracy of result depends on basis set. Changing the method and basis set will lead to large difference in total energy of molecule and we can never compare the result of analysis using different basis set.
(2) What is the purpose of carrying out a frequency analysis?
Answer: As frequency analysis is the second derivative of the potential energy surface, positive frequency indicates minimum of energy and negative frequency indicates a transition state, which can be used to confirm the previous optimisation is correct. Besides, it provides information of vibrational frequency and IR spectrum that can be compared between molecules.
(3) What do the "Low frequencies" represent?
Answer: If low frequencies do not contain negative values then we know the optimisation succeed.
Molecular Orbital Analysis of BH3
1. D-space Link
The D-space link of the MO analysis is [4].
2. MO Visualisation
| MO number | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| GaussView computed MO |  |
 |
 |
 |
 |
 |
 |

|
| Symmetry label | a1' | a1' | e' | e' | a2' | a1' | e' | e' |
4. Questions
(1) Are there any significant differences between the real and LCAO MOs?
Answer: The real MOs are 3D which can be visualised using GuassView in different angles, while LCAO MOs are predicted in 2D. But overall they match well.
(2) What does this say about the accuracy and usefulness of qualitative MO theory?
Answer: MO theory predicts LCAO MOs well, which can be used as a good indication of the real MOs.
NH3 Analysis
NH3 Optimisation
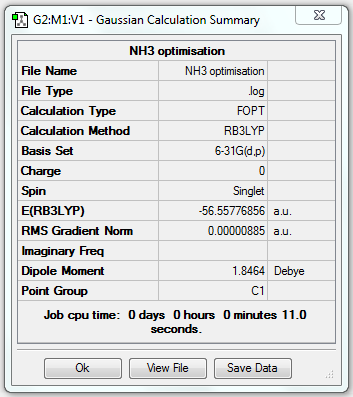
1. Optimising NH3 Using Basis Set 6-31G(d,p)
2. File Link
The file of the optimisation is here.
3. Geometric Information
N-H bond length = 1.01797Å, H-N-H bond angle = 105.741o.
5. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629730D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
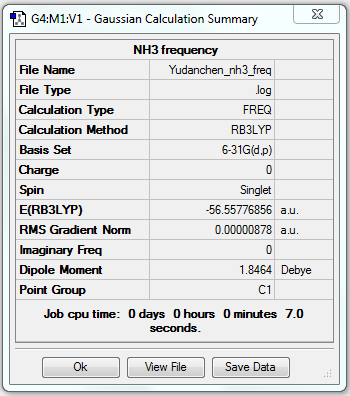
Frequency Analysis of NH3
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -30.8045 -0.0008 -0.0005 0.0013 20.2188 28.2150 Low frequencies --- 1089.5530 1694.1235 1694.1861
MO Analysis of NH3
1. File Link
The file of the MO analysis is here.
NBO Analysis of NH3
1. Charge Distribution Image of NH3
The color range is set from -1.131 to 1.131.
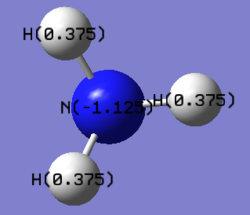
2. Specific NBO Charges of NH3
The color range is set from -1.131 to 1.131.
3. "Real" Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756
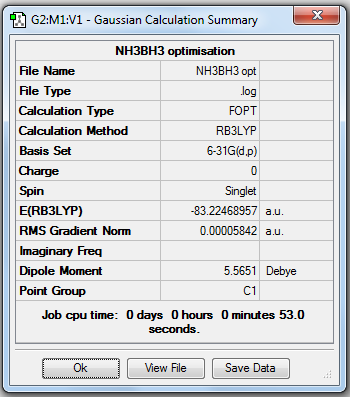
NH3BH3 Analysis
NH3BH3 Optimisation
1. Optimising NH3BH3 Using Basis Set 6-31G(d,p)

2. File Link
The file of the optimisation is here.
4. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000124 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000660 0.001800 YES
RMS Displacement 0.000304 0.001200 YES
Predicted change in Energy=-1.649831D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.21 -DE/DX = -0.0001 !
! R2 R(2,8) 1.21 -DE/DX = -0.0001 !
! R3 R(3,8) 1.21 -DE/DX = -0.0001 !
! R4 R(4,7) 1.0186 -DE/DX = -0.0001 !
! R5 R(5,7) 1.0186 -DE/DX = -0.0001 !
! R6 R(6,7) 1.0186 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(4,7,5) 107.8698 -DE/DX = 0.0 !
! A2 A(4,7,6) 107.8683 -DE/DX = 0.0 !
! A3 A(4,7,8) 111.0271 -DE/DX = 0.0 !
! A4 A(5,7,6) 107.87 -DE/DX = 0.0 !
! A5 A(5,7,8) 111.0319 -DE/DX = 0.0 !
! A6 A(6,7,8) 111.0285 -DE/DX = 0.0 !
! A7 A(1,8,2) 113.8754 -DE/DX = 0.0 !
! A8 A(1,8,3) 113.8742 -DE/DX = 0.0 !
! A9 A(1,8,7) 104.5931 -DE/DX = 0.0 !
! A10 A(2,8,3) 113.8743 -DE/DX = 0.0 !
! A11 A(2,8,7) 104.5962 -DE/DX = 0.0 !
! A12 A(3,8,7) 104.6001 -DE/DX = 0.0 !
! D1 D(4,7,8,1) 179.998 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -60.0028 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 59.9984 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -60.001 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 59.9983 -DE/DX = 0.0 !
! D6 D(5,7,8,3) 179.9994 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 60.0012 -DE/DX = 0.0 !
! D8 D(6,7,8,2) 180.0005 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -59.9984 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
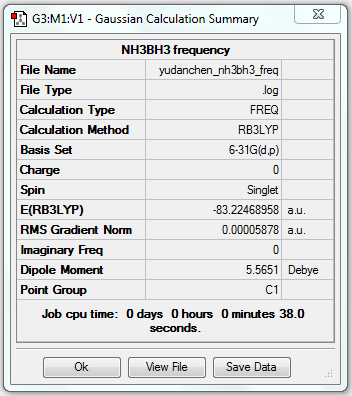
Frequency Analysis of NH3BH3
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -0.0006 -0.0003 0.0013 14.8232 19.6226 45.9567 Low frequencies --- 267.5738 632.2147 639.3062
Dissociation Energy of NH3BH3
| E(NH3) | E(BH3) | E(NH3BH3) |
|---|---|---|
| -56.55776856a.u. | -26.61532363a.u. | -83.22468957a.u. |
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]=-83.22468957-[(-56.55776856)+(-26.61532363)]=-0.05159738a.u.=-135.46892119kJ/mol
Week 2 Mini Project: Investigating Aromaticity
Benzene Analysis
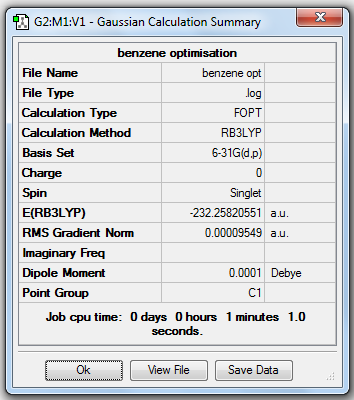
Benzene Optimisation (6-31G(d,p))
1. Optimising Benzene Using Basis Set 6-31G(d,p)

2. D-space Link
The D-space of the optimisation is [5].
4. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000212 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000991 0.001800 YES
RMS Displacement 0.000315 0.001200 YES
Predicted change in Energy=-5.157454D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.086 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.086 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9972 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9949 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0079 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0079 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9881 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.004 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9948 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0086 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9966 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9972 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9934 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0094 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0083 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0014 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9904 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9946 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0106 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9948 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0023 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.01 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0013 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.007 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0117 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.9914 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0036 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0005 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0062 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0059 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9969 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0028 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0051 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0058 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0055 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0054 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0082 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
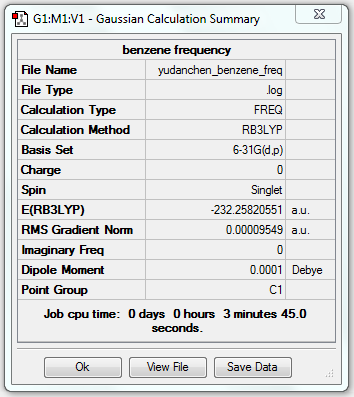
Frequency Analysis of Benzene
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -14.2788 -11.5868 -9.6527 -0.0009 0.0007 0.0004 Low frequencies --- 413.7971 414.4697 620.8545
MO Analysis of Benzene
1. D-space Link
The D-space of the MO analysis is [6].
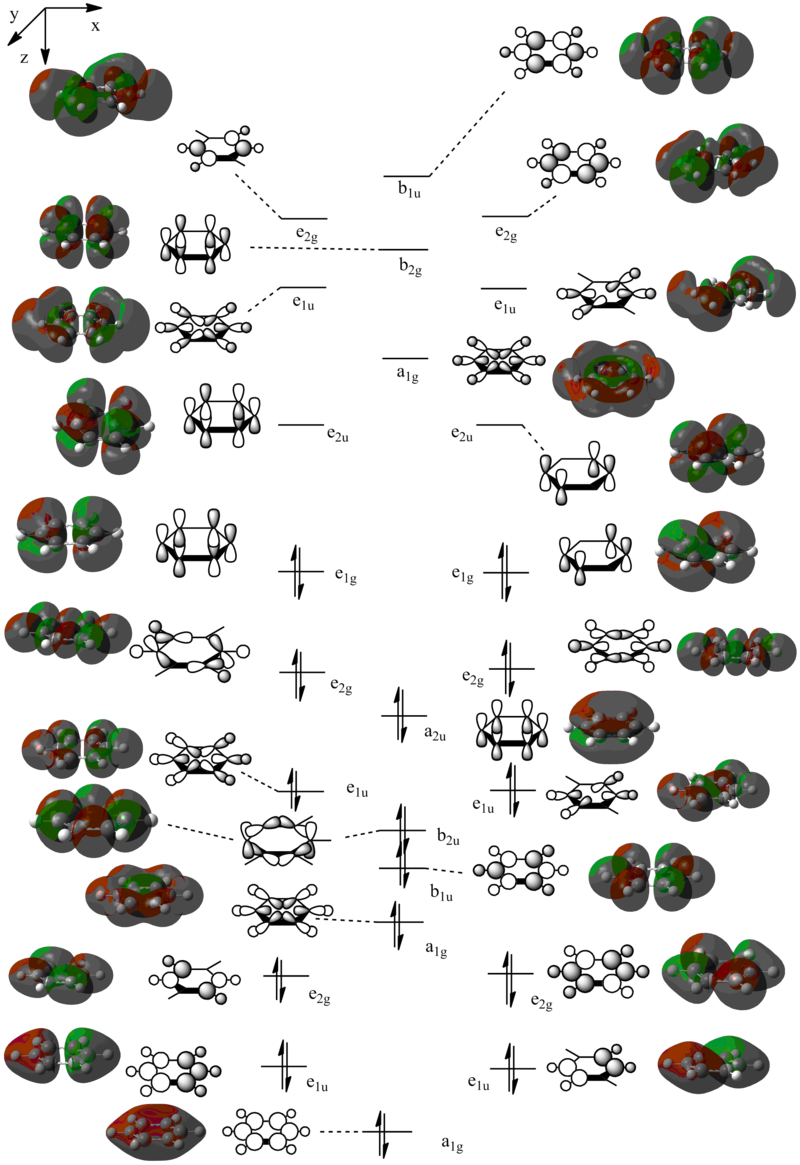
3. MO Diagram
NBO Analysis of Benzene
1. Charge Distribution Image of Benzene
The color range is set from -0.239 to 0.239.
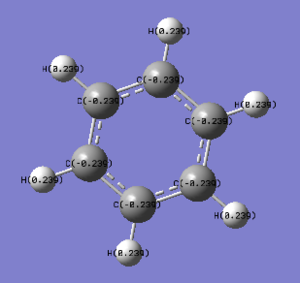
2. Specific NBO Charges of Benzene
The color range is set from -0.239 to 0.239.
3. "Real" Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.23852 1.99910 4.22611 0.01331 6.23852
C 2 -0.23855 1.99910 4.22613 0.01331 6.23855
C 3 -0.23854 1.99910 4.22613 0.01331 6.23854
C 4 -0.23852 1.99910 4.22611 0.01331 6.23852
C 5 -0.23855 1.99910 4.22613 0.01331 6.23855
C 6 -0.23854 1.99910 4.22613 0.01331 6.23854
H 7 0.23854 0.00000 0.76003 0.00144 0.76146
H 8 0.23853 0.00000 0.76003 0.00144 0.76147
H 9 0.23854 0.00000 0.76002 0.00144 0.76146
H 10 0.23854 0.00000 0.76003 0.00144 0.76146
H 11 0.23853 0.00000 0.76003 0.00144 0.76147
H 12 0.23854 0.00000 0.76002 0.00144 0.76146
=======================================================================
* Total * 0.00000 11.99462 29.91690 0.08847 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98096) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.7508
-0.0048 -0.2875 -0.0354 -0.0001 0.0000
0.0092 0.0000 0.0000 0.0138 -0.0109
( 50.00%) 0.7071* C 2 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.7645
0.0260 0.2488 -0.0245 0.0000 0.0000
0.0116 0.0000 0.0000 0.0119 -0.0109
2. (1.98097) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.1669
0.0342 0.7864 0.0103 0.0000 0.0000
0.0045 0.0000 0.0000 -0.0160 -0.0109
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.1263
0.0282 -0.7940 -0.0219 0.0000 0.0000
0.0073 0.0000 0.0000 -0.0149 -0.0109
3. (1.66532) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 -0.0066 0.0183 0.0000 0.0000
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 -0.0128 -0.0147 0.0000 0.0000
4. (1.98305) BD ( 1) C 1 - H 7
( 62.04%) 0.7876* C 1 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.6377
-0.0111 -0.5450 0.0095 0.0000 0.0000
-0.0164 0.0000 0.0000 0.0026 -0.0105
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0173 0.0148 0.0000
5. (1.98098) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.21%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.6244
-0.0331 0.5064 -0.0135 0.0000 0.0000
-0.0165 0.0000 0.0000 0.0011 -0.0109
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.5977
-0.0082 -0.5377 -0.0348 -0.0001 0.0000
-0.0161 0.0000 0.0000 0.0041 -0.0109
6. (1.66534) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 -0.0064 0.0184 0.0000 0.0000
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0001
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 0.0192 -0.0034 0.0000 0.0000
7. (1.98305) BD ( 1) C 2 - H 8
( 62.04%) 0.7876* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5436 -0.0126 0.0010 0.1529
-0.0027 0.8248 -0.0144 0.0000 0.0000
-0.0060 0.0000 0.0000 0.0155 0.0105
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0042 -0.0224 0.0000
8. (1.98096) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.1264
-0.0282 0.7940 0.0218 0.0000 0.0000
0.0073 0.0000 0.0000 -0.0149 -0.0109
( 50.00%) 0.7071* C 4 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.1669
-0.0342 -0.7865 -0.0103 0.0000 0.0000
0.0045 0.0000 0.0000 -0.0160 -0.0109
9. (1.98305) BD ( 1) C 3 - H 9
( 62.04%) 0.7876* C 3 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.7908
-0.0138 0.2798 -0.0049 -0.0001 0.0000
-0.0105 0.0000 0.0000 -0.0129 0.0105
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0215 -0.0076 0.0000
10. (1.98098) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.7508
0.0048 0.2875 0.0354 -0.0001 0.0000
0.0092 0.0000 0.0000 0.0138 -0.0109
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.7645
-0.0260 -0.2487 0.0245 0.0000 0.0000
0.0116 0.0000 0.0000 0.0119 -0.0109
11. (1.66533) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0001 0.0000 0.9997 -0.0133
0.0000 0.0191 -0.0037 0.0000 0.0000
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0133
0.0000 -0.0125 -0.0149 0.0000 0.0000
12. (1.98305) BD ( 1) C 4 - H 10
( 62.04%) 0.7876* C 4 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.6378
-0.0111 -0.5449 0.0095 0.0000 0.0000
0.0164 0.0000 0.0000 -0.0026 0.0105
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0173 0.0148 0.0000
13. (1.98096) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.6244
0.0330 -0.5064 0.0135 0.0001 0.0000
-0.0165 0.0000 0.0000 0.0011 -0.0109
( 50.00%) 0.7071* C 6 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.5978
0.0082 0.5377 0.0347 -0.0001 0.0000
-0.0161 0.0000 0.0000 0.0041 -0.0109
14. (1.98305) BD ( 1) C 5 - H 11
( 62.04%) 0.7876* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5436 0.0126 -0.0010 0.1531
-0.0027 0.8248 -0.0143 0.0000 0.0000
0.0060 0.0000 0.0000 -0.0155 -0.0105
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0042 -0.0224 0.0000
15. (1.98305) BD ( 1) C 6 - H 12
( 62.04%) 0.7876* C 6 s( 29.58%)p 2.38( 70.38%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.7907
-0.0138 0.2800 -0.0049 0.0000 0.0000
0.0105 0.0000 0.0000 0.0129 -0.0105
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0215 -0.0076 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98096 -0.68184 110(g),107(g),120(v),114(v)
43(v),73(v),109(g),112(g)
72(v),42(v)
2. BD ( 1) C 1 - C 6 1.98097 -0.68199 118(g),106(g),119(v),112(v)
63(v),33(v),120(g),109(g)
32(v),62(v)
3. BD ( 2) C 1 - C 6 1.66532 -0.23794 116(v),111(v),35(v),65(v)
4. BD ( 1) C 1 - H 7 1.98305 -0.51236 118(v),110(v),72(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98098 -0.68202 106(g),113(g),117(v),109(v)
23(v),53(v),114(g),112(g)
52(v),22(v)
6. BD ( 2) C 2 - C 3 1.66534 -0.23795 116(v),108(v),25(v),55(v)
7. BD ( 1) C 2 - H 8 1.98305 -0.51233 113(v),107(v),42(v),22(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98096 -0.68183 110(g),115(g),112(v),119(v)
33(v),63(v),114(g),117(g)
62(v),32(v)
9. BD ( 1) C 3 - H 9 1.98305 -0.51236 106(v),115(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - C 5 1.98098 -0.68200 118(g),113(g),114(v),120(v)
73(v),43(v),117(g),119(g)
42(v),72(v)
11. BD ( 2) C 4 - C 5 1.66533 -0.23794 108(v),111(v),45(v),75(v)
12. BD ( 1) C 4 - H 10 1.98305 -0.51236 118(v),110(v),62(v),42(v)
115(g),113(g)
13. BD ( 1) C 5 - C 6 1.98096 -0.68186 115(g),107(g),109(v),117(v)
53(v),23(v),119(g),120(g)
22(v),52(v)
14. BD ( 1) C 5 - H 11 1.98305 -0.51233 113(v),107(v),52(v),72(v)
115(g),118(g)
15. BD ( 1) C 6 - H 12 1.98305 -0.51236 106(v),115(v),22(v),62(v)
107(g),118(g)
16. CR ( 1) C 1 1.99911 -10.04057 73(v),33(v),110(v),118(v)
120(v),112(v)
17. CR ( 1) C 2 1.99911 -10.04056 43(v),23(v),113(v),107(v)
114(v),109(v)
18. CR ( 1) C 3 1.99911 -10.04056 33(v),53(v),106(v),115(v)
112(v),117(v)
19. CR ( 1) C 4 1.99911 -10.04057 63(v),43(v),118(v),110(v)
119(v),114(v)
20. CR ( 1) C 5 1.99911 -10.04056 53(v),73(v),113(v),107(v)
117(v),120(v)
21. CR ( 1) C 6 1.99911 -10.04056 23(v),63(v),115(v),106(v)
109(v),119(v)
Boratabenzene Analysis
Boratabenzene Optimisation (6-31G(d,p))
1. Optimising Boratabenzene Using Basis Set 6-31G(d,p)

2. File Link
The file of the optimisation is here.
4. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000878 0.001800 YES
RMS Displacement 0.000326 0.001200 YES
Predicted change in Energy=-6.589451D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4053 -DE/DX = -0.0001 !
! R2 R(1,5) 1.3989 -DE/DX = 0.0 !
! R3 R(1,6) 1.0968 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,7) 1.0916 -DE/DX = -0.0001 !
! R6 R(3,4) 1.3989 -DE/DX = 0.0 !
! R7 R(3,8) 1.0968 -DE/DX = 0.0001 !
! R8 R(4,9) 1.097 -DE/DX = -0.0001 !
! R9 R(4,12) 1.5137 -DE/DX = 0.0001 !
! R10 R(5,11) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5138 -DE/DX = 0.0001 !
! R12 R(10,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,5) 122.138 -DE/DX = 0.0001 !
! A2 A(2,1,6) 117.4354 -DE/DX = 0.0 !
! A3 A(5,1,6) 120.4266 -DE/DX = -0.0002 !
! A4 A(1,2,3) 120.4508 -DE/DX = -0.0001 !
! A5 A(1,2,7) 119.7734 -DE/DX = 0.0001 !
! A6 A(3,2,7) 119.7758 -DE/DX = 0.0001 !
! A7 A(2,3,4) 122.1395 -DE/DX = 0.0001 !
! A8 A(2,3,8) 117.4371 -DE/DX = 0.0 !
! A9 A(4,3,8) 120.4234 -DE/DX = -0.0002 !
! A10 A(3,4,9) 115.9493 -DE/DX = 0.0001 !
! A11 A(3,4,12) 120.0806 -DE/DX = -0.0001 !
! A12 A(9,4,12) 123.9701 -DE/DX = -0.0001 !
! A13 A(1,5,11) 115.9535 -DE/DX = 0.0001 !
! A14 A(1,5,12) 120.0812 -DE/DX = -0.0001 !
! A15 A(11,5,12) 123.9654 -DE/DX = -0.0001 !
! A16 A(4,12,5) 115.1098 -DE/DX = 0.0 !
! A17 A(4,12,10) 122.4482 -DE/DX = 0.0 !
! A18 A(5,12,10) 122.4419 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0057 -DE/DX = 0.0 !
! D2 D(5,1,2,7) 180.0027 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0038 -DE/DX = 0.0 !
! D4 D(6,1,2,7) 0.0008 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0018 -DE/DX = 0.0 !
! D6 D(2,1,5,12) -0.001 -DE/DX = 0.0 !
! D7 D(6,1,5,11) 0.0002 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.001 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0074 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0016 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.0044 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0014 -DE/DX = 0.0 !
! D13 D(2,3,4,9) 180.0049 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0042 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0011 -DE/DX = 0.0 !
! D16 D(8,3,4,12) -180.0018 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0005 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -180.0 -DE/DX = 0.0 !
! D19 D(9,4,12,5) -180.0003 -DE/DX = 0.0 !
! D20 D(9,4,12,10) -0.0008 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.002 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.0015 -DE/DX = 0.0 !
! D23 D(11,5,12,4) 179.9989 -DE/DX = 0.0 !
! D24 D(11,5,12,10) -0.0006 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
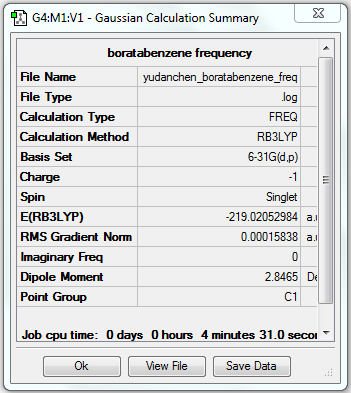
Frequency Analysis of Boratabenzene
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -13.1275 -0.0008 -0.0001 0.0006 15.0447 18.1653 Low frequencies --- 371.3454 404.2334 565.2534
MO Analysis of Boratabenzene
1. File Link
The file of the MO analysis is here.
NBO Analysis of Boratabenzene
1. Charge Distribution Image of Boratabenzene
The color range is set from -0.588 to 0.588.
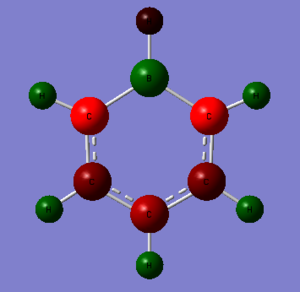
2. Specific NBO Charges of Boratabenzene
The color range is set from -0.588 to 0.588.

3. "Real" Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.25043 1.99910 4.23720 0.01412 6.25043
C 2 -0.33984 1.99907 4.32693 0.01384 6.33984
C 3 -0.25041 1.99910 4.23719 0.01412 6.25041
C 4 -0.58795 1.99901 4.57715 0.01178 6.58795
C 5 -0.58790 1.99901 4.57711 0.01178 6.58790
H 6 0.17906 0.00000 0.81832 0.00262 0.82094
H 7 0.18564 0.00000 0.81237 0.00200 0.81436
H 8 0.17906 0.00000 0.81832 0.00262 0.82094
H 9 0.18380 0.00000 0.81402 0.00218 0.81620
H 10 -0.09642 0.00000 1.09588 0.00054 1.09642
H 11 0.18380 0.00000 0.81402 0.00218 0.81620
B 12 0.20160 1.99906 2.78774 0.01160 4.79840
=======================================================================
* Total * -1.00000 11.99436 29.91625 0.08939 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.97970) BD ( 1) C 1 - C 2
( 49.96%) 0.7068* C 1 s( 35.50%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6874
-0.0034 -0.4135 -0.0325 0.0000 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7062
0.0327 0.3754 -0.0141 0.0000 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
2. (1.98270) BD ( 1) C 1 - C 5
( 50.77%) 0.7125* C 1 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0573
0.0311 0.7869 0.0164 0.0000 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 5 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0025
0.0269 -0.8201 -0.0353 0.0000 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
3. (1.76867) BD ( 2) C 1 - C 5
( 48.13%) 0.6938* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
4. (1.98570) BD ( 1) C 1 - H 6
( 59.32%) 0.7702* C 1 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7228
-0.0089 -0.4562 0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 6 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
5. (1.97971) BD ( 1) C 2 - C 3
( 50.04%) 0.7074* C 2 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7063
-0.0327 0.3752 -0.0141 0.0000 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 3 s( 35.51%)p 1.82( 64.45%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6875
0.0034 -0.4133 -0.0325 0.0000 0.0000
-0.0146 0.0000 0.0000 0.0081 -0.0107
6. (1.98507) BD ( 1) C 2 - H 7
( 59.44%) 0.7710* C 2 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 -0.0001
0.0000 0.8469 -0.0076 0.0000 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
7. (1.98270) BD ( 1) C 3 - C 4
( 50.77%) 0.7125* C 3 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0575
-0.0311 0.7869 0.0164 0.0000 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 4 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 0.0027
-0.0269 -0.8202 -0.0353 0.0000 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
8. (1.76862) BD ( 2) C 3 - C 4
( 48.13%) 0.6937* C 3 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
9. (1.98570) BD ( 1) C 3 - H 8
( 59.32%) 0.7702* C 3 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
0.0003 -0.5182 -0.0133 0.0012 0.7227
-0.0089 0.4563 -0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
10. (1.98420) BD ( 1) C 4 - H 9
( 59.41%) 0.7708* C 4 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7907
0.0003 0.3469 0.0088 0.0000 0.0000
-0.0111 0.0000 0.0000 0.0150 -0.0119
( 40.59%) 0.6371* H 9 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
11. (1.96997) BD ( 1) C 4 - B 12
( 66.70%) 0.8167* C 4 s( 42.04%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 0.6482 0.0158 0.0012 0.6115
-0.0293 0.4524 0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7056
-0.0393 -0.4071 0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
12. (1.98420) BD ( 1) C 5 - H 11
( 59.41%) 0.7708* C 5 s( 25.39%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7906
-0.0003 0.3471 0.0088 0.0000 0.0000
0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
13. (1.96996) BD ( 1) C 5 - B 12
( 66.70%) 0.8167* C 5 s( 42.03%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6481 -0.0158 -0.0012 0.6117
-0.0293 -0.4522 -0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7057
-0.0393 0.4070 -0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
14. (1.98604) BD ( 1) H 10 - B 12
( 55.09%) 0.7422* H 10 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 -0.0001
0.0000 0.8172 -0.0016 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
15. (1.99910) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
1.0000 0.0003 0.0000 0.0000 0.0002
0.0000 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.97970 -0.46973 108(g),111(g),115(v),118(v)
44(v),63(v),112(g),110(g)
2. BD ( 1) C 1 - C 5 1.98270 -0.46498 119(g),107(g),112(v),110(g)
34(v),120(v),118(g),98(v)
33(v)
3. BD ( 2) C 1 - C 5 1.76867 -0.02908 21(v),22(v),35(v),100(v)
109(g)
4. BD ( 1) C 1 - H 6 1.98570 -0.31413 111(v),119(v),33(v),63(v)
108(g)
5. BD ( 1) C 2 - C 3 1.97971 -0.46976 113(g),107(g),110(v),116(v)
24(v),53(v),112(g),115(g)
6. BD ( 1) C 2 - H 7 1.98507 -0.31744 113(v),108(v),43(v),23(v)
111(g),107(g)
7. BD ( 1) C 3 - C 4 1.98270 -0.46495 117(g),111(g),112(v),115(g)
34(v),120(v),116(g),98(v)
33(v)
8. BD ( 2) C 3 - C 4 1.76862 -0.02907 21(v),22(v),35(v),100(v)
114(g)
9. BD ( 1) C 3 - H 8 1.98570 -0.31412 107(v),117(v),33(v),53(v)
113(g)
10. BD ( 1) C 4 - H 9 1.98420 -0.28848 111(v),117(g),43(v),119(v)
97(v),113(g)
11. BD ( 1) C 4 - B 12 1.96997 -0.31779 113(g),115(v),118(v),116(g)
44(v),43(v),85(v),64(v)
119(g)
12. BD ( 1) C 5 - H 11 1.98420 -0.28849 107(v),119(g),23(v),117(v)
97(v),108(g)
13. BD ( 1) C 5 - B 12 1.96996 -0.31776 108(g),110(v),116(v),118(g)
24(v),23(v),93(v),54(v)
117(g)
14. BD ( 1) H 10 - B 12 1.98604 -0.17253 113(v),108(v),53(v),63(v)
15. CR ( 1) C 1 1.99910 -9.83478 64(v),34(v),119(v),112(v)
111(v),118(v)
16. CR ( 1) C 2 1.99907 -9.82827 44(v),24(v),113(v),108(v)
115(v),110(v),27(v),47(v)
43(v),23(v)
17. CR ( 1) C 3 1.99910 -9.83478 54(v),34(v),117(v),112(v)
107(v),116(v)
18. CR ( 1) C 4 1.99902 -9.79407 44(v),98(v),117(g),111(v)
115(v),97(v)
19. CR ( 1) C 5 1.99902 -9.79408 24(v),98(v),119(g),107(v)
110(v),97(v)
20. CR ( 1) B 12 1.99907 -6.36942 116(v),118(v),113(v),108(v)
53(v),63(v)
21. LP ( 1) C 2 1.14688 0.09689 114(v),109(v),35(g),46(v)
26(v),45(v),25(v)
22. LP*( 1) B 12 0.57262 0.22267 114(v),109(v),100(g),57(v)
67(v)
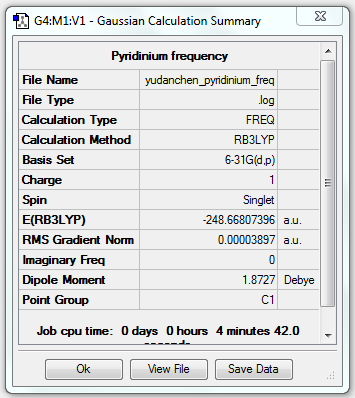
Pyridinium Analysis
Pyridinium Optimisation (6-31G(d,p))
1. Optimising Pyridinium Using Basis Set 6-31G(d,p)
2. File Link
The file of the optimisation is here.
4. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000065 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000826 0.001800 YES
RMS Displacement 0.000176 0.001200 YES
Predicted change in Energy=-6.972574D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,5) 1.3838 -DE/DX = 0.0 !
! R3 R(1,6) 1.0835 -DE/DX = 0.0 !
! R4 R(2,3) 1.3988 -DE/DX = 0.0 !
! R5 R(2,7) 1.0852 -DE/DX = 0.0 !
! R6 R(3,4) 1.3839 -DE/DX = 0.0 !
! R7 R(3,8) 1.0835 -DE/DX = 0.0 !
! R8 R(4,9) 1.0832 -DE/DX = 0.0 !
! R9 R(4,12) 1.3523 -DE/DX = 0.0001 !
! R10 R(5,11) 1.0832 -DE/DX = 0.0 !
! R11 R(5,12) 1.3524 -DE/DX = 0.0 !
! R12 R(10,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 119.0827 -DE/DX = 0.0 !
! A2 A(2,1,6) 121.496 -DE/DX = -0.0001 !
! A3 A(5,1,6) 119.4213 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0549 -DE/DX = 0.0 !
! A5 A(1,2,7) 119.9711 -DE/DX = 0.0 !
! A6 A(3,2,7) 119.974 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.082 -DE/DX = 0.0 !
! A8 A(2,3,8) 121.4988 -DE/DX = -0.0001 !
! A9 A(4,3,8) 119.4192 -DE/DX = 0.0001 !
! A10 A(3,4,9) 123.9297 -DE/DX = 0.0 !
! A11 A(3,4,12) 119.2363 -DE/DX = 0.0 !
! A12 A(9,4,12) 116.834 -DE/DX = 0.0 !
! A13 A(1,5,11) 123.9327 -DE/DX = 0.0 !
! A14 A(1,5,12) 119.2354 -DE/DX = 0.0 !
! A15 A(11,5,12) 116.8319 -DE/DX = 0.0 !
! A16 A(4,12,5) 123.3088 -DE/DX = 0.0 !
! A17 A(4,12,10) 118.3463 -DE/DX = 0.0 !
! A18 A(5,12,10) 118.345 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0006 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0023 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0015 -DE/DX = 0.0 !
! D4 D(6,1,2,7) -0.0014 -DE/DX = 0.0 !
! D5 D(2,1,5,11) -180.0005 -DE/DX = 0.0 !
! D6 D(2,1,5,12) 0.0021 -DE/DX = 0.0 !
! D7 D(6,1,5,11) -0.0014 -DE/DX = 0.0 !
! D8 D(6,1,5,12) 180.0012 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.0024 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0018 -DE/DX = 0.0 !
! D11 D(7,2,3,4) 180.0005 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0012 -DE/DX = 0.0 !
! D13 D(2,3,4,9) 180.0002 -DE/DX = 0.0 !
! D14 D(2,3,4,12) 0.0015 -DE/DX = 0.0 !
! D15 D(8,3,4,9) -0.0004 -DE/DX = 0.0 !
! D16 D(8,3,4,12) 180.0008 -DE/DX = 0.0 !
! D17 D(3,4,12,5) 0.0014 -DE/DX = 0.0 !
! D18 D(3,4,12,10) -180.001 -DE/DX = 0.0 !
! D19 D(9,4,12,5) 180.0025 -DE/DX = 0.0 !
! D20 D(9,4,12,10) 0.0001 -DE/DX = 0.0 !
! D21 D(1,5,12,4) -0.0032 -DE/DX = 0.0 !
! D22 D(1,5,12,10) -180.0008 -DE/DX = 0.0 !
! D23 D(11,5,12,4) -180.0007 -DE/DX = 0.0 !
! D24 D(11,5,12,10) 0.0016 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis of Pyridinium
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -7.2115 -0.0012 -0.0010 -0.0004 17.3444 18.5499 Low frequencies --- 392.4572 404.0614 620.4717
MO Analysis of Pyridinium
1. File Link
The file of the MO analysis is here.
NBO Analysis of Pyridinium
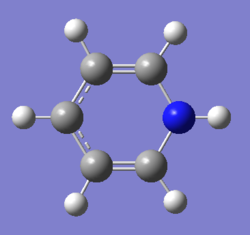
1. Charge Distribution Image of Pyridinium
The color range is set from -0.483 to 0.483.
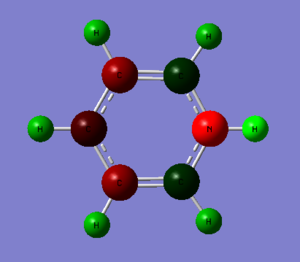
2. Specific NBO Charges of Pyridinium
The color range is set from -0.483 to 0.483.

3. "Real" Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.24103 1.99912 4.22860 0.01331 6.24103
C 2 -0.12241 1.99913 4.10941 0.01386 6.12241
C 3 -0.24104 1.99912 4.22860 0.01331 6.24104
C 4 0.07100 1.99918 3.91066 0.01916 5.92900
C 5 0.07098 1.99918 3.91068 0.01916 5.92902
H 6 0.29718 0.00000 0.70179 0.00103 0.70282
H 7 0.29170 0.00000 0.70718 0.00113 0.70830
H 8 0.29718 0.00000 0.70179 0.00103 0.70282
H 9 0.28493 0.00000 0.71397 0.00110 0.71507
H 10 0.48279 0.00000 0.51475 0.00246 0.51721
H 11 0.28493 0.00000 0.71397 0.00110 0.71507
N 12 -0.47622 1.99937 5.46756 0.00929 7.47622
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98249) BD ( 1) C 1 - C 2
( 50.26%) 0.7089* C 1 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4185
-0.0371 -0.6897 0.0068 0.0000 0.0000
0.0122 0.0000 0.0000 -0.0118 -0.0115
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3935
-0.0234 0.7063 0.0290 0.0000 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
2. (1.98297) BD ( 1) C 1 - C 5
( 49.58%) 0.7042* C 1 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 -0.0009 0.0320 0.0000 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 5 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 0.0142 0.0331 0.0000 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
3. (1.61445) BD ( 2) C 1 - C 5
( 52.23%) 0.7227* C 1 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0068
0.0000 0.0191 -0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9995 -0.0175
0.0000 -0.0196 -0.0153 0.0000 0.0000
4. (1.97822) BD ( 1) C 1 - H 6
( 64.83%) 0.8052* C 1 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 -0.3988
0.0072 0.7229 -0.0181 0.0000 0.0000
-0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 6 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 0.0116 -0.0208 0.0000
5. (1.98249) BD ( 1) C 2 - C 3
( 49.74%) 0.7053* C 2 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3934
-0.0234 -0.7064 -0.0290 0.0000 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 3 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4184
-0.0371 0.6898 -0.0068 0.0000 0.0000
-0.0122 0.0000 0.0000 -0.0118 -0.0115
6. (1.54879) BD ( 2) C 2 - C 3
( 45.73%) 0.6762* C 2 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0036
0.0000 0.0241 -0.0101 0.0000 0.0000
( 54.27%) 0.7367* C 3 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0080
0.0000 0.0086 0.0228 0.0000 0.0000
7. (1.98141) BD ( 1) C 2 - H 7
( 64.64%) 0.8040* C 2 s( 31.07%)p 2.22( 68.89%)d 0.00( 0.03%)
0.0003 -0.5573 -0.0131 0.0007 0.8298
-0.0198 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0153 0.0101
( 35.36%) 0.5947* H 7 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0242 0.0000 0.0000
8. (1.98297) BD ( 1) C 3 - C 4
( 49.58%) 0.7042* C 3 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 0.0007 -0.0320 0.0000 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 4 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 -0.0140 -0.0331 0.0000 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
9. (1.97822) BD ( 1) C 3 - H 8
( 64.83%) 0.8052* C 3 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 0.3989
-0.0072 0.7228 -0.0181 0.0000 0.0000
-0.0109 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 8 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 -0.0116 -0.0208 0.0000
10. (1.98154) BD ( 1) C 4 - H 9
( 64.26%) 0.8016* C 4 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 -0.4693
0.0193 0.6666 -0.0183 0.0000 0.0000
0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 9 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 0.0128 -0.0209 0.0000
11. (1.98861) BD ( 1) C 4 - N 12
( 36.68%) 0.6057* C 4 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4043
0.0563 0.7416 0.0277 0.0000 0.0000
0.0252 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 -0.3660
0.0187 -0.7069 -0.0132 0.0000 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0115
12. (1.82447) BD ( 2) C 4 - N 12
( 28.55%) 0.5343* C 4 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9991 0.0132
0.0000 0.0102 0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9999 0.0036
0.0000 -0.0128 -0.0077 0.0000 0.0000
13. (1.98154) BD ( 1) C 5 - H 11
( 64.26%) 0.8016* C 5 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 0.4694
-0.0193 0.6664 -0.0183 0.0000 0.0000
0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 11 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0128 -0.0209 0.0000
14. (1.98861) BD ( 1) C 5 - N 12
( 36.68%) 0.6057* C 5 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4042
0.0563 -0.7417 -0.0277 0.0000 0.0000
-0.0252 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 -0.3658
0.0187 0.7069 0.0132 0.0000 0.0000
-0.0107 0.0000 0.0000 -0.0059 -0.0115
15. (1.98630) BD ( 1) H 10 - N 12
( 25.41%) 0.5041* H 10 s( 99.88%)p 0.00( 0.12%)
0.9994 -0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.82%)p 2.73( 73.16%)d 0.00( 0.02%)
-0.0002 0.5178 0.0066 -0.0013 0.8553
-0.0091 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0115 -0.0106
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98249 -0.90377 118(v),114(v),107(g),63(v)
110(g),109(g),43(v),42(v)
112(g)
2. BD ( 1) C 1 - C 5 1.98297 -0.92650 120(v),112(v),106(g),33(v)
118(g),97(v),109(g),32(v)
119(g),96(v)
3. BD ( 2) C 1 - C 5 1.61445 -0.46666 111(v),117(v),99(v),108(g)
37(v)
4. BD ( 1) C 1 - H 6 1.97822 -0.71855 119(v),110(v),32(v),62(v)
106(g),107(g)
5. BD ( 1) C 2 - C 3 1.98249 -0.90379 115(v),109(v),113(g),54(v)
106(g),114(g),23(v),22(v)
112(g)
6. BD ( 2) C 2 - C 3 1.54879 -0.44891 117(v),108(v),56(v),26(v)
7. BD ( 1) C 2 - H 7 1.98141 -0.71806 113(v),107(v),22(v),42(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98297 -0.92648 120(v),112(v),110(g),33(v)
115(g),97(v),114(g),32(v)
116(g),96(v)
9. BD ( 1) C 3 - H 8 1.97822 -0.71855 116(v),106(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - H 9 1.98154 -0.75115 119(v),110(v),42(v),113(g)
96(v)
11. BD ( 1) C 4 - N 12 1.98861 -1.06562 119(g),62(v),114(v),43(v)
118(v),63(v),113(g),120(g)
12. BD ( 2) C 4 - N 12 1.82447 -0.56812 108(v),111(v),64(v),46(v)
13. BD ( 1) C 5 - H 11 1.98154 -0.75115 116(v),106(v),22(v),107(g)
96(v)
14. BD ( 1) C 5 - N 12 1.98861 -1.06559 116(g),52(v),109(v),23(v)
115(v),54(v),107(g),120(g)
15. BD ( 1) H 10 - N 12 1.98630 -0.89229 113(v),107(v),52(v),62(v)
16. CR ( 1) C 1 1.99913 -10.26482 33(v),63(v),62(v),118(v)
110(v),72(v),112(v),119(v)
17. CR ( 1) C 2 1.99914 -10.27397 43(v),23(v),107(v),113(v)
114(v),109(v),76(v)
18. CR ( 1) C 3 1.99913 -10.26482 33(v),54(v),52(v),115(v)
106(v),80(v),112(v),116(v)
19. CR ( 1) C 4 1.99918 -10.32333 43(v),119(v),120(v),110(v)
113(g),97(v),84(v),114(v)
20. CR ( 1) C 5 1.99918 -10.32332 23(v),116(v),120(v),106(v)
107(g),97(v),92(v),109(v)
21. CR ( 1) N 12 1.99937 -14.46220 54(v),63(v)
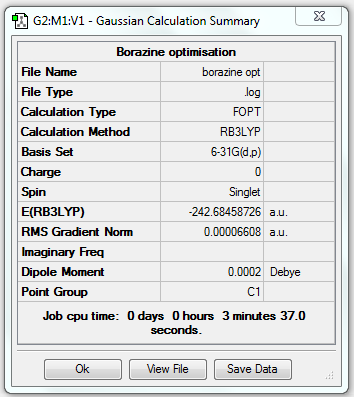
Borazine Analysis
Borazine Optimisation (6-31G(d,p))
1. Optimising Borazine Using Basis Set 6-31G(d,p)
2. File Link
The file of the optimisation is here.
4. "Real" Output
Item Value Threshold Converged?
Maximum Force 0.000093 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000341 0.001800 YES
RMS Displacement 0.000096 0.001200 YES
Predicted change in Energy=-1.028631D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,9) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,12) 1.0097 -DE/DX = 0.0 !
! R3 R(3,8) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,10) 1.0097 -DE/DX = 0.0 !
! R5 R(5,7) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,11) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4306 -DE/DX = -0.0001 !
! R8 R(7,11) 1.4307 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4307 -DE/DX = -0.0001 !
! R10 R(8,12) 1.4307 -DE/DX = 0.0 !
! R11 R(9,11) 1.4306 -DE/DX = 0.0 !
! R12 R(9,12) 1.4307 -DE/DX = 0.0 !
! A1 A(5,7,10) 121.4436 -DE/DX = 0.0 !
! A2 A(5,7,11) 121.4373 -DE/DX = 0.0 !
! A3 A(10,7,11) 117.119 -DE/DX = 0.0001 !
! A4 A(3,8,10) 121.4337 -DE/DX = 0.0 !
! A5 A(3,8,12) 121.4348 -DE/DX = 0.0 !
! A6 A(10,8,12) 117.1314 -DE/DX = 0.0 !
! A7 A(1,9,11) 121.4414 -DE/DX = 0.0 !
! A8 A(1,9,12) 121.4399 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1187 -DE/DX = 0.0 !
! A10 A(4,10,7) 118.567 -DE/DX = 0.0 !
! A11 A(4,10,8) 118.5613 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.8717 -DE/DX = 0.0 !
! A13 A(6,11,7) 118.5563 -DE/DX = 0.0 !
! A14 A(6,11,9) 118.5584 -DE/DX = 0.0 !
! A15 A(7,11,9) 122.8853 -DE/DX = 0.0 !
! A16 A(2,12,8) 118.5639 -DE/DX = 0.0 !
! A17 A(2,12,9) 118.5622 -DE/DX = 0.0 !
! A18 A(8,12,9) 122.8739 -DE/DX = 0.0 !
! D1 D(5,7,10,4) 0.0043 -DE/DX = 0.0 !
! D2 D(5,7,10,8) -179.9997 -DE/DX = 0.0 !
! D3 D(11,7,10,4) -179.9957 -DE/DX = 0.0 !
! D4 D(11,7,10,8) 0.0002 -DE/DX = 0.0 !
! D5 D(5,7,11,6) 0.0003 -DE/DX = 0.0 !
! D6 D(5,7,11,9) 179.9967 -DE/DX = 0.0 !
! D7 D(10,7,11,6) -179.9996 -DE/DX = 0.0 !
! D8 D(10,7,11,9) -0.0033 -DE/DX = 0.0 !
! D9 D(3,8,10,4) 0.0005 -DE/DX = 0.0 !
! D10 D(3,8,10,7) -179.9954 -DE/DX = 0.0 !
! D11 D(12,8,10,4) -179.9991 -DE/DX = 0.0 !
! D12 D(12,8,10,7) 0.005 -DE/DX = 0.0 !
! D13 D(3,8,12,2) 0.0034 -DE/DX = 0.0 !
! D14 D(3,8,12,9) 179.9928 -DE/DX = 0.0 !
! D15 D(10,8,12,2) -179.997 -DE/DX = 0.0 !
! D16 D(10,8,12,9) -0.0075 -DE/DX = 0.0 !
! D17 D(1,9,11,6) -0.0025 -DE/DX = 0.0 !
! D18 D(1,9,11,7) -179.9989 -DE/DX = 0.0 !
! D19 D(12,9,11,6) 179.9973 -DE/DX = 0.0 !
! D20 D(12,9,11,7) 0.0009 -DE/DX = 0.0 !
! D21 D(1,9,12,2) -0.0061 -DE/DX = 0.0 !
! D22 D(1,9,12,8) -179.9955 -DE/DX = 0.0 !
! D23 D(11,9,12,2) 179.9942 -DE/DX = 0.0 !
! D24 D(11,9,12,8) 0.0048 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
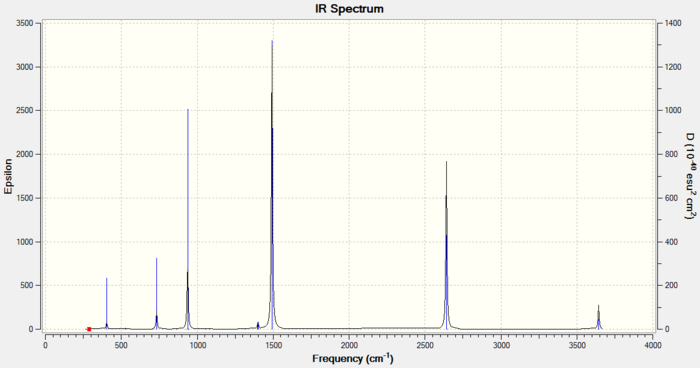
Frequency Analysis of Borazine
1. File Link
The file of the frequency analysis is here.
3. "Real" Output
Low frequencies --- -17.2356 -10.7561 -6.7538 -0.0009 0.0004 0.0006 Low frequencies --- 288.8544 289.6660 404.1667
MO Analysis of Borazine
1. File Link
The file of the MO analysis is here.
NBO Analysis of Borazine
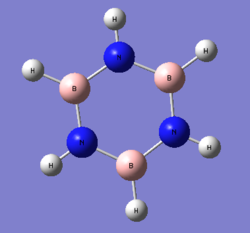
1. Charge Distribution Image of Borazine
The color range is set from -1.102 to 1.102.
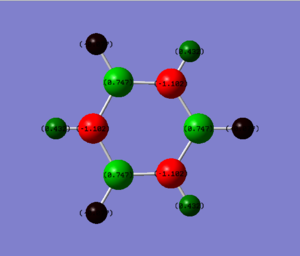
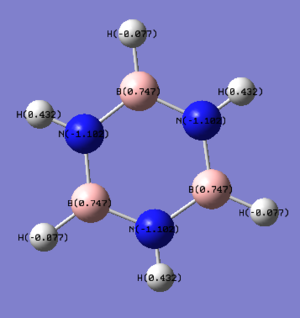
2. Specific NBO Charges of Borazine
The color range is set from -1.102 to 1.102.
3. "Real" Output
******************************Gaussian NBO Version 3.1******************************
N A T U R A L A T O M I C O R B I T A L A N D
N A T U R A L B O N D O R B I T A L A N A L Y S I S
******************************Gaussian NBO Version 3.1******************************
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43198 0.00000 0.56573 0.00228 0.56802
H 3 -0.07655 0.00000 1.07586 0.00069 1.07655
H 4 0.43198 0.00000 0.56574 0.00228 0.56802
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43198 0.00000 0.56574 0.00228 0.56802
B 7 0.74700 1.99917 2.23862 0.01520 4.25300
B 8 0.74696 1.99917 2.23866 0.01521 4.25304
B 9 0.74695 1.99917 2.23868 0.01521 4.25305
N 10 -1.10241 1.99943 6.09820 0.00478 8.10241
N 11 -1.10241 1.99943 6.09821 0.00478 8.10241
N 12 -1.10240 1.99943 6.09819 0.00478 8.10240
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06889 42.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98670) BD ( 1) H 1 - B 9
( 54.03%) 0.7351* H 1 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 -0.0176 0.0077 0.0000
( 45.97%) 0.6780* B 9 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 0.7241
-0.0247 -0.3156 0.0107 0.0000 0.0000
-0.0173 0.0000 0.0000 0.0161 -0.0098
2. (1.98495) BD ( 1) H 2 - N 12
( 28.08%) 0.5299* H 2 s( 99.91%)p 0.00( 0.09%)
0.9996 -0.0010 -0.0238 -0.0175 0.0000
( 71.92%) 0.8481* N 12 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
-0.0002 0.4776 -0.0114 0.0006 0.7064
0.0105 0.5218 0.0078 0.0000 0.0000
0.0116 0.0000 0.0000 0.0036 -0.0119
3. (1.98670) BD ( 1) H 3 - B 8
( 54.03%) 0.7351* H 3 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0022 -0.0191 0.0000
( 45.97%) 0.6780* B 8 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.0887
0.0030 0.7849 -0.0267 0.0000 0.0000
-0.0053 0.0000 0.0000 -0.0230 -0.0098
4. (1.98495) BD ( 1) H 4 - N 10
( 28.08%) 0.5299* H 4 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 -0.0271 0.0118 0.0000
( 71.92%) 0.8481* N 10 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 0.8051
0.0120 -0.3510 -0.0052 0.0000 0.0000
0.0089 0.0000 0.0000 -0.0083 0.0119
5. (1.98670) BD ( 1) H 5 - B 7
( 54.03%) 0.7351* H 5 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0154 0.0114 0.0000
( 45.97%) 0.6780* B 7 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.6353
0.0216 -0.4693 0.0160 0.0000 0.0000
0.0226 0.0000 0.0000 0.0069 -0.0098
6. (1.98495) BD ( 1) H 6 - N 11
( 28.08%) 0.5299* H 6 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 0.0033 -0.0293 0.0000
( 71.92%) 0.8481* N 11 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 -0.0987
-0.0015 0.8727 0.0130 0.0000 0.0000
0.0027 0.0000 0.0000 0.0119 0.0119
7. (1.98438) BD ( 1) B 7 - N 10
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.0705
0.0408 -0.8226 -0.0410 0.0000 0.0000
0.0045 0.0000 0.0000 0.0450 0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.0272
0.0145 0.7832 -0.0065 0.0000 0.0000
0.0007 0.0000 0.0000 0.0072 0.0085
8. (1.82090) BD ( 2) B 7 - N 10
( 11.79%) 0.3433* B 7 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 0.0027 0.0613 0.0000 0.0000
( 88.21%) 0.9392* N 10 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 0.0040 -0.0023 0.0000 0.0000
9. (1.98438) BD ( 1) B 7 - N 11
( 23.53%) 0.4851* B 7 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.7656
-0.0272 0.3091 0.0510 0.0000 0.0000
0.0290 0.0000 0.0000 -0.0347 0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7406
-0.0019 -0.2563 0.0158 0.0000 0.0000
0.0046 0.0000 0.0000 -0.0056 0.0085
10. (1.98438) BD ( 1) B 8 - N 10
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.6506
-0.0578 -0.5084 0.0020 0.0000 0.0000
0.0445 0.0000 0.0000 0.0078 -0.0206
( 76.47%) 0.8745* N 10 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.5923
-0.0146 0.5132 0.0063 0.0000 0.0000
0.0071 0.0000 0.0000 0.0012 -0.0085
11. (1.98438) BD ( 1) B 8 - N 12
( 23.53%) 0.4851* B 8 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.7476
-0.0559 0.3503 -0.0148 0.0001 0.0000
0.0367 0.0000 0.0000 -0.0264 0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.6919
-0.0129 -0.3680 -0.0094 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0042 0.0085
12. (1.82091) BD ( 2) B 8 - N 12
( 11.79%) 0.3433* B 8 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9976 -0.0315
0.0000 0.0518 -0.0330 0.0000 0.0000
( 88.21%) 0.9392* N 12 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 -0.0040 -0.0023 0.0000 0.0000
13. (1.98438) BD ( 1) B 9 - N 11
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.6772
-0.0151 -0.4723 -0.0558 0.0000 0.0000
0.0412 0.0000 0.0000 0.0186 -0.0206
( 76.47%) 0.8745* N 11 s( 38.55%)p 1.59( 61.43%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.6646
0.0017 0.4152 -0.0158 0.0000 0.0000
0.0065 0.0000 0.0000 0.0030 -0.0085
14. (1.82093) BD ( 2) B 9 - N 11
( 11.79%) 0.3433* B 9 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9976 -0.0315
0.0000 -0.0545 -0.0283 0.0000 0.0000
( 88.21%) 0.9392* N 11 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 1.0000 -0.0003
0.0000 0.0000 0.0046 0.0000 0.0000
15. (1.98438) BD ( 1) B 9 - N 12
( 23.53%) 0.4851* B 9 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.1150
-0.0306 -0.8176 -0.0491 0.0000 0.0000
0.0155 0.0000 0.0000 0.0425 0.0206
( 76.47%) 0.8745* N 12 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.1484
-0.0127 0.7695 -0.0095 0.0000 0.0000
0.0025 0.0000 0.0000 0.0067 0.0085
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H6B3N3)
1. BD ( 1) H 1 - B 9 1.98670 -0.40394 114(v),116(v),86(v),96(v)
2. BD ( 1) H 2 - N 12 1.98495 -0.61482 115(v),118(v),116(g),120(g)
56(v),66(v)
3. BD ( 1) H 3 - B 8 1.98670 -0.40392 120(v),112(v),76(v),96(v)
4. BD ( 1) H 4 - N 10 1.98495 -0.61483 116(v),114(v),112(g),115(g)
46(v),56(v)
5. BD ( 1) H 5 - B 7 1.98670 -0.40394 115(v),118(v),76(v),86(v)
6. BD ( 1) H 6 - N 11 1.98495 -0.61480 120(v),112(v),118(g),114(g)
46(v),66(v)
7. BD ( 1) B 7 - N 10 1.98438 -0.68871 115(g),111(v),109(g),108(v)
57(v),116(v)
8. BD ( 2) B 7 - N 10 1.82090 -0.27139 117(v),62(v),58(v),35(v)
113(g)
9. BD ( 1) B 7 - N 11 1.98438 -0.68868 118(g),109(v),111(g),106(v)
67(v),120(v)
10. BD ( 1) B 8 - N 10 1.98438 -0.68869 112(g),107(v),109(g),110(v)
47(v),114(v)
11. BD ( 1) B 8 - N 12 1.98438 -0.68871 120(g),109(v),107(g),106(v)
67(v),118(v)
12. BD ( 2) B 8 - N 12 1.82091 -0.27139 119(v),72(v),68(v),27(v)
117(g)
13. BD ( 1) B 9 - N 11 1.98438 -0.68873 114(g),107(v),111(g),110(v)
47(v),112(v)
14. BD ( 2) B 9 - N 11 1.82093 -0.27140 113(v),52(v),48(v),43(v)
119(g)
15. BD ( 1) B 9 - N 12 1.98438 -0.68871 116(g),111(v),107(g),108(v)
57(v),115(v)
16. CR ( 1) B 7 1.99917 -6.65247 115(v),118(v),109(v),111(v)
17. CR ( 1) B 8 1.99917 -6.65246 120(v),112(v),107(v),109(v)
18. CR ( 1) B 9 1.99917 -6.65245 114(v),116(v),107(v),111(v)
19. CR ( 1) N 10 1.99943 -14.13096 47(v),57(v),112(g),115(g)
20. CR ( 1) N 11 1.99943 -14.13097 67(v),47(v),118(g),114(g)
21. CR ( 1) N 12 1.99943 -14.13097 67(v),57(v),120(g),116(g)
Result and Discussion
Aromaticity
All the four molecules fulfill Huckel's Rule, which is if a cyclic, planar molecule has 4n+2 pi electrons (in which n=0,1,2,3,...) then it is considered as aromatic. From the MOs we can see strongly delocolised pi electron clouds along the ring. And each MO only contains 2 electrons giving an agreement to 4n+2=2 where n=0.
Charge Distribution Comparison
In benzene, the C atoms and H atoms have the same magnitude of charge number, which is probably because of its highly symmetric structure. As the B atom is highly electropositive, it tends to donate electron density onto the ring leading to a negative charge on boratabenzene, and thus the charge of C atoms and H atoms of boratabenzene becomes more negative than that of benzene. Similarly, the N atom makes the charge of C atoms and H atoms more positive due to its relatively high electronegativity, resulting in a positive charge on pyridinium. For borazine, the alternative N atom and B atom leads to a large charge difference of the surrounding atoms, but overall the dipole moments cancel out and thus give a charge of zero.
MO Comparison
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| MO 21(HOMO) |  |
 |
 |

|
| MO 20 | 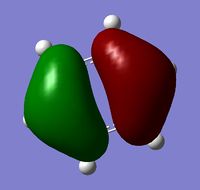 |
 |
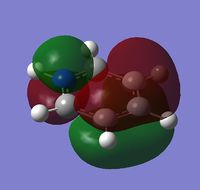 |
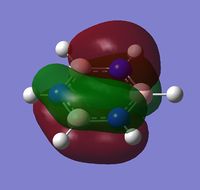
|
| MO 19 | 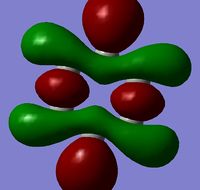 |
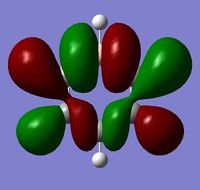 |
 |
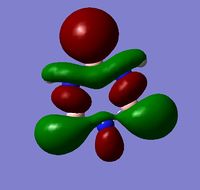
|
The MOs show that the p orbital bonding is similar for the four molecules. Benzene has the highest symmetry. When hetero-atoms are introduced into this highly symmetric aromatic system, the electronegativity difference between the surrounding atoms increases, leading to unequal electron sharing, therefore the symmetry of MOs is reduced for Boratabenzene, Pyridinium and borazine.
| Molecule | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| MO 21(HOMO) energy | -0.24691 | 0.01094 | -0.47885 | -0.27590 |
| MO 20 energy | -0.24692 | -0.03493 | -0.50847 | -0.27591 |
| MO 19 energy | -0.33960 | -0.08387 | -0.57432 | -0.31993 |
Boratabenzene has a higher MO energy than benzene, as the electropositive B atom shifts the energy level of one of the LCAOs up, leading to larger energy gap between the two LCAOs and thus destablising the resulting MO. Similarly, pyridinium has a lower MO energy than benzene due to the electronegative N atom. For boranzine, the overall dipole moment cancel out, therefore the hetero-atoms only have a small effect on MO energy.
Effect of substitutions on MO diagram
When one or more C atoms are substituted by hetero-atoms, the changes in electronegativity shift the energy level of LCAOs up or down depending on the relative electronegativity of the hetero-atoms. For example, in the case of pypiridinium, when a C atom is substituted by a N atom, the energy level of one of the LCAOs is lowered and thus the energy levels of the two LCAOs are closer and have a greater stablisation in the bonding MO.
References
- ↑ JULIUS GLASER and GEORG JOHANSSON, On the Structures of the Hydrated Thallium(III) Ion and its Bromide Complexes in Aqueous Solution, Acta Chemica Scandinavica, A, 36, 1982, 125.