Rep:Mod:yrtaxol2
EXPERIMENT 1C
Name: Yong R. Tan
CID: 00703262
Part 1
Hydrogenation of Tricyclo[5.2.1.02,6]deca-3,8-diene (or commonly know as dicyclopentadiene).
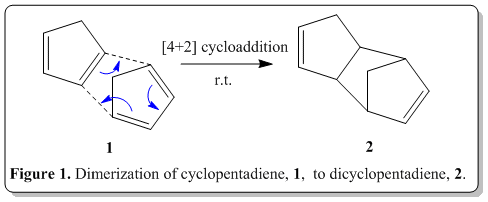
Tricyclo[5.2.1.02,6]deca-3,8-diene is readily available inexpensively in coal and tar. Upon reaching, its boiling point (170o) it would depolymerize back to its monomer, cyclopentadiene, which has boiling point of 41°, and can be distilled off to obtain the pure cyclopentadiene. This process is commonly known as cracking. When left at room temperature, cyclopentadiene eventually dimerizes back to dicyclopentadiene. The polymerization can be determined when there is a change in specific gravity of the compound after some days(specific gravity of cyclopentadiene: 0.805; of dicyclopentadiene, 0.976).[1]
The polymerization process, is actually a Nobel prize winning reaction, namely Diels-Alder reaction. Via the classical Diels-Alder reaction, two cyclopentadiene molecules, 1 would dimerize (as shown in Figure 1) to yield dicyclopentadiene, 2, which is a facile and spontaneous [4+2] cycloaddition reaction at room temperature. The product can either be an exo, 3 or endo, 4 product, as shown in Figure 2.
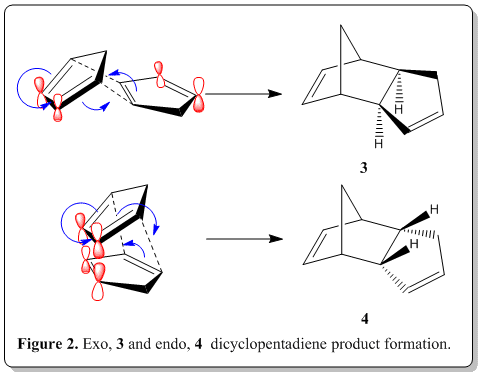
From experimental results reported in the literature[2], the thermal dimerization gives exclusively the endo dicyclopentadiene, 4 only and does not have any exo dicyclopentadiene, 3 formed. It was known that the endo product is the kinetic product while the exo product is the thromodynamic product. Also, from the studies of Alder and Stein[3], they indicated that the endo-form had the higher energy content.
Computational analysis of the Tricyclo[5.2.1.02,6]deca-3,8-diene was executed with the Avogadro program. The endo and exo dicyclopentadiene geometries were optimized by means of the MMFF94(s) force field option. The alogrithm used was the conjugate gradient option as this evaluates the value of the diagonal terms only while setting all the others to zero. Though it is slower than the steepest descent option, but it assuages the vacillation when reaches less 'steep' gradient where in the steepest descent option, it is very fast but suffer from the vacillation upon reaching less 'steep' gradient. The present results are consistent with the experimental observations and experimental results reported in the literature[3] as mentioned earlier.
| Types of Total Energy | Exo, 3 / kcal mol-1 | Endo, 4 / kcal mol-1 | ||||||
| TOTAL BOND STRETCHING ENERGY | 3.54302 | 3.46760 | ||||||
| TOTAL ANGLE BENDING ENERGY | 30.77273 | 33.19065 | ||||||
| TOTAL STRETCH BENDING ENERGY | -2.04139 | -2.08217 | ||||||
| TOTAL TORSIONAL ENERGY | -2.73105 | -2.94978 | ||||||
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.01486 | 0.02198 | ||||||
| TOTAL VAN DER WAALS ENERGY | 12.80155 | 12.35764 | ||||||
| TOTAL ELECTROSTATIC ENERGY | 13.01372 | 14.18480 | ||||||
| TOTAL ENERGY | 55.37344 | 58.19070 | ||||||
| Model |
|
|
From our calculations shown in the table above, the total energy of 3 (exo product, 55.37344 kcal mol-1) is smaller than of the 4 (endo product, 58.19070 kcal mol-1). This concur with the results reported by Alder and Stein [3] where the endo product is higher energetic than of the exo product. From the calculation, the endo product is identified as the kinetic product (being higher in energy) while the exo product is identified as the thermodynamically stable product (bearing lower energy).
From the experimental results[2], the endo dicyclopentadiene, 4 is the sole product but from our calculation, 4 is the conformation with higher energy than of 3(in thermodynamic sense). Yet this is plausible because despite it being the higher energy content, we should inspect the mechanism of the Diels-Alder reaction which lead to the product where stereoselectivity is controlled by stereoelectronic factors. As shown in Figure 2, when the reaction proceeds to form 4, we are able to see that there are bonding interactions between the frontier orbitals (labelled red). The symmetry of the orbitals labelled in red are the same and thus able to have such bonding interactions where in 3, it does not have such advantage. This interaction resulted in this preferred orientation during the transition state. As soon as the cycloaddition is complete, the product had the same orientation as during the transition state as the stereochemistry was already defined, leaving the product being exclusively endo only. The endo product is energetically higher which is a kinetic product and thus we could conclude that the Diels-Alder reaction of dimerization ofcyclopentadiene is under kinetic control, leaving the exo product which is thermodynamically more stable being unformed at all.
Other than that, the two main calculated energies for 3 and 4 that dominated the difference in energy, was the Total Angle Bending Energy (EAB)and Total Electrostatic Energy(EE). Both the energies are higher in 4 than of 3. The EAB for 4 is 3 kcal/mol higher than of 3. This could be attributed to the fact that 4 is the endo product, and the ring would be forced to 'bend' inwards, stressing the bicyclo rings to be bending in a more strained conformation, consequent to the higher EAB. Also, the EE of 4 is 1 kcal/mol higher than of 3. As explained earlier, the resultant of the straining of the increases the Coulombic potential within the molecule which resulted in the computed values for EE.
In short, as this reaction is under kinetic control, despite 4 being the highly energetic configuration, it is formed predominantly.
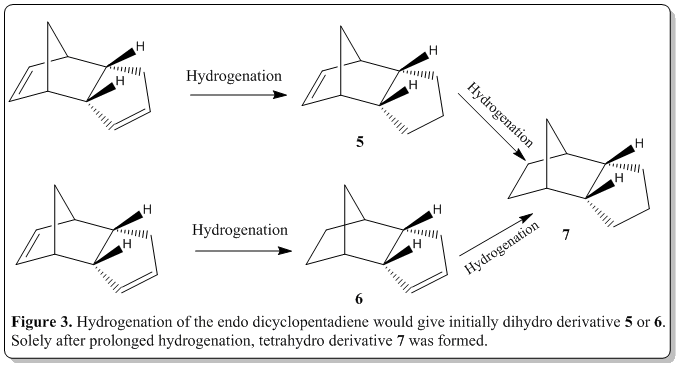
Also, from the endo dicyclopentadiene, when hydrogenation was performed, the dimer would yield, initially one of the dihydro derivative 5 or 6. Then with prolonged hydrogenation, only will the tetrahydro derivative 7 be formed. The reaction scheme is as shown in Figure 3. From the literature, the results was that 6 would be formed predominantly then formation of 7. [4] Would the calculations following the discussion show something that could rationalize this observation?
Computational analysis of 5 and 6 were executed with the Avogadro program. They were optimized by means of the MMFF94(s) force field option. The alogrithm used was the conjugate gradient option as this evaluates the value of the diagonal terms only while setting all the others to zero. Though it is slower than the steepest descent option, but it assuages the vacillation when reaches less 'steep' gradient where in the steepest descent option, it is very fast but suffer from the vacillation upon reaching less 'steep' gradient. The results and conformation of the computed values are summarized below:
| Types of Total Energy | Endo, 4 / kcal mol-1 | Dihydro, 5 / kcal mol-1 | Dihydro, 6 / kcal mol-1 | Tetrahydro, 7 / kcal mol-1 | ||||||||||||
| TOTAL BOND STRETCHING ENERGY | 3.46760 | 5.28510 | 2.82306 | 2.68756 | ||||||||||||
| TOTAL ANGLE BENDING ENERGY | 33.19065 | 36.54346 | 24.68552 | 22.64378 | ||||||||||||
| TOTAL STRETCH BENDING ENERGY | -2.08217 | -1.83542 | -1.65716 | -1.51556 | ||||||||||||
| TOTAL TORSIONAL ENERGY | -2.94978 | -3.70581 | -0.37813 | 3.30168 | ||||||||||||
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.02198 | 0.04191 | 0.00028 | 0.00000 | ||||||||||||
| TOTAL VAN DER WAALS ENERGY | 12.35764 | 16.41094 | 10.63690 | 10.89720 | ||||||||||||
| TOTAL ELECTROSTATIC ENERGY | 14.18480 | 5.14499 | 5.14702 | 0.00000 | ||||||||||||
| TOTAL ENERGY | 58.19070 | 57.88517 | 41.25749 | 38.01467 | ||||||||||||
| Model |
|
|
|
|
From the calculation being done, it was computed that the hydrogenation product 5 is higher in steric energy than 6 by 16.6 kcal/mol. As from the literature[4] , the ease of forming hydrogenation product 6 is higher than of 5. Combine with the literature value and our calculation, we could postulate that the hydrogenation is under thermodynamic control where the product stability factor predominates in this reaction.
The energies that resulted in the difference in energy were the total bond stretching energy (EBS), total angle bending energy (EAB), total torsional energy(ET), and total Van der Waals energy(EVDW).The total electrostatic energy of both 5 and 6 are almost similar. In general, EBS, EAB, ET, and EVDW are all higher in 5 than of 6. The energies listed are higher in 5 than of 6 was due to product 5 still bears the alkene in a five-membered ring but the ring was strained due to it being part of the bicyclic system. On the other hand, the product 6 has the alkene in a five-membered ring but the ring was not part of the strained bicyclic ring and thus it has lower overall strain energy. As the hhydrogenation is under thermodynamic control, 6 is formed first then only with prolonged hydrogenation, 7 is formed. As anticipated, product 7 is lower in energy than 5 and 6. as the whole system does not have any alkene functional groups that strains the whole system and engendered the lowering in energy.
Atropisomerism of a large ring ketone intermediate in one synthesis of the anti-cancer drug Taxol and insights on bridgehead alkene reactivities.
(i) Atropisomerism
 [5]
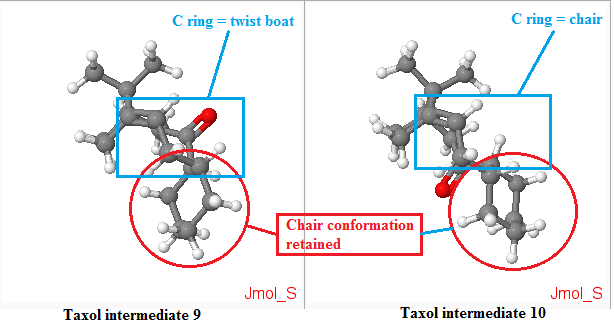
[5] From the literature[5], compound 8 undergoes oxy-Cope rearrangement to either compound 9 or compound 10. The difference between compound 9 and 10 are the positions of the carbonyl (Colored red in Figure 4.), either pointing up (C ring = twist boat) or down (C-ring = chair). As reported by Elmore[5], having the carbonyl group pointing up in compound 9 would lock the fused cyclohexane (ring C) into an energitically expensive twist-boat conformation. When the carbonyl group is pointing down, the situation is alleviated where chair-like C ring of 10, non-bonded transannular interactions are heightened within the B ring core of such structure.
Computational analysis of 9 and 10 were executed with the Avogadro program. They were optimized by means of the MMFF94(s) force field option. The alogrithm used was the conjugate gradient option as this evaluates the value of the diagonal terms only while setting all the others to zero. Though it is slower than the steepest descent option, but it assuages the vacillation when reaches less 'steep' gradient where in the steepest descent option, it is very fast but suffer from the vacillation upon reaching less 'steep' gradient. The results and conformation of the computed values are summarized below:
| Types of Total Energy | Taxol intermediate, 9 / kcal mol-1 | Taxol intermediate, 10 / kcal mol-1 | ||||||
| TOTAL BOND STRETCHING ENERGY | 7.71700 | 7.60144 | ||||||
| TOTAL ANGLE BENDING ENERGY | 28.30969 | 18.79205 | ||||||
| TOTAL STRETCH BENDING ENERGY | -0.06819 | -0.14341 | ||||||
| TOTAL TORSIONAL ENERGY | 0.08236 | 0.21194 | ||||||
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.96319 | 0.84732 | ||||||
| TOTAL VAN DER WAALS ENERGY | 33.24990 | 33.29756 | ||||||
| TOTAL ELECTROSTATIC ENERGY | 0.29397 | -0.05295 | ||||||
| TOTAL ENERGY | 70.54792 | 60.55395 | ||||||
| Model |
|
|
From the table above, the Taxol intermediate 9 is higher in energy than Taxol intermediate 10, and the difference is by about 10 kcal mol-1 higher. The total angle bending energy appeared to be the energy that distinguished the energy difference of 9 and 10. As the cyclohexane ring had the chair conformation retained (as shown in figure below), the main difference is in the strain of the ring as labelled in the blue box in figure below. In the Taxol intermediate 10, the ring is in chair conformation while in 9, it is in twist boat conformation which is the source of the hike in energy for conformation 9. The carbonyl group pointing up or down locks the ring in certain conformation and for this case, when it is pointing down (Taxol intermediate 10), it locks the whole system in much lower energy conformation.
(ii)Stability of bridgehead alkenes
Other than that, it was worth noting that for both the intermediates, there was a bridgehead alkene present. Historically, Bredt [6]
Computational analysis of 9, product of hydrogenation of 9, 10 and product of hydrogenation of 10 were executed with the Avogadro program. They were optimized by means of the MMFF94(s) force field option. The alogrithm used was the conjugate gradient option as this evaluates the value of the diagonal terms only while setting all the others to zero. Though it is slower than the steepest descent option, but it assuages the vacillation when reaches less 'steep' gradient where in the steepest descent option, it is very fast but suffer from the vacillation upon reaching less 'steep' gradient. The results and conformation of the computed values are summarized below:
| Types of Total Energy | Taxol intermediate, 9 / kcal mol-1 | Hydrogenation of 9 / kcal | Taxol intermediate, 10 / kcal mol-1 | Hydrogenation of 10 / kcal | ||||||||||||
| TOTAL BOND STRETCHING ENERGY | 7.71700 | 12.29091 | 7.60144 | 7.30881 | ||||||||||||
| TOTAL ANGLE BENDING ENERGY | 28.30969 | 48.45715 | 18.79205 | 25.92577 | ||||||||||||
| TOTAL STRETCH BENDING ENERGY | -0.06819 | 1.21082 | -0.14341 | 0.20145 | ||||||||||||
| TOTAL TORSIONAL ENERGY | 0.08236 | 5.68683 | 0.21194 | 6.79650 | ||||||||||||
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.96319 | 0.10975 | 0.84732 | 0.09989 | ||||||||||||
| TOTAL VAN DER WAALS ENERGY | 33.24990 | 41.97101 | 33.29756 | 33.41936 | ||||||||||||
| TOTAL ELECTROSTATIC ENERGY | 0.29397 | 0.00000 | -0.05295 | 0.00000 | ||||||||||||
| TOTAL ENERGY | 70.54792 | 109.72648 | 60.55395 | 73.75179 | ||||||||||||
| Model |
|
|
|
|
From table above, it was computed that both the product of hydrogenation of the 9 and 10 yielded in product of much higher in energy where for the hydrogenation of 9, the product became about 30 kcal/mol higher in energy! Meanwhile, hydrogenation of 10 becomes 13 kcal/mol higher in energy. Upon detailed inspection on the data for hydrogenatio of 10, it was found that the total angle bending energy (EAB) and total torsional energy (ET) were the two contributing energies that raises the overall strain energy of the hydrogenated product of 10. On the other hand, for hydrogenation of 9, the total bond stretching energy (EBS), total angle bending energy (EAB), total torsional energy (ET), and total Van der Waals energy (EVDW) were the responsible in the massive increase in the strain energy of the hydrogenated product of 9.
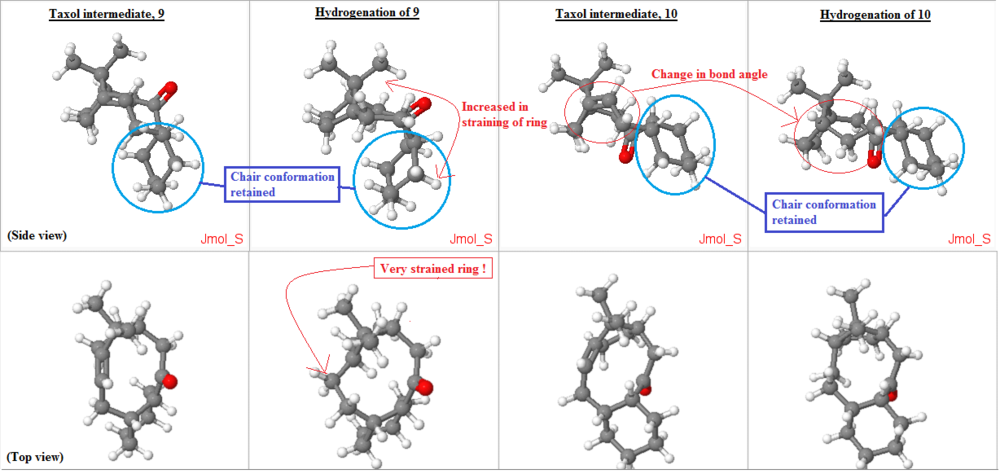
From figure above, all the structures were optimized with their cyclohexane ring in chair conformation to ensure that the overall structure would be in its lowest conformation possible. (circled and labelled in blue). In the hydrogenation of 10, when the sp2 carbons in the alkene was converted to sp3 carbons, there was a change in bond angle of 120 deg. to 109.5 deg. this reduction in bond angle (as depicted in the figure above with red circle) increases the ring strain of the molecule as the ring need to be existing with smaller bond angle. This is very unfavorable. Thus, resulting in overall increase in the energy of the molecule, making the hydrogenation of the alkene at such bridgehead become unfavorable thermodynamically.
Meanwhile, for the hydrogenation of 9, there was a massive increase in energy of the product! It shows that it is even less favorable to hydrogenate such bridgehead alkene. As from the top view figure, on the "Hydrogenation of 9" column, the change in ring shape induced a even strained ring as can be seen from the figure above the point where the ring have to squeeze a corner out from the original ring due to the change in the sp2 carbon to sp3 carbon after hydrogenation. Similar to the previous case, there was a change in bond angle of 120 deg. to 109.5 deg. this reduction in bond angle (as depicted in the figure above with red circle) increases the ring strain of the molecule as the ring need to be existing with smaller bond angle. This is very unfavorable as well. Plus, from the side view, one can spot that the ring has to curve inwards (as shown in the figure above, on the side view, 'Hydrogenation of 9' column), straining the ring even more.
In short, the hydrogenation of these bridgehead alkenes would yield product which is thermodynamically high energy products and thus very unfavourable. These alkenes react very slowly and are also coined as hyperstable alkenes too.
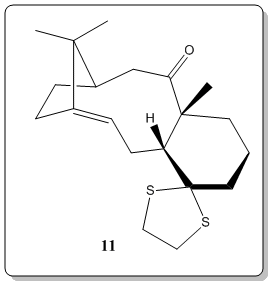
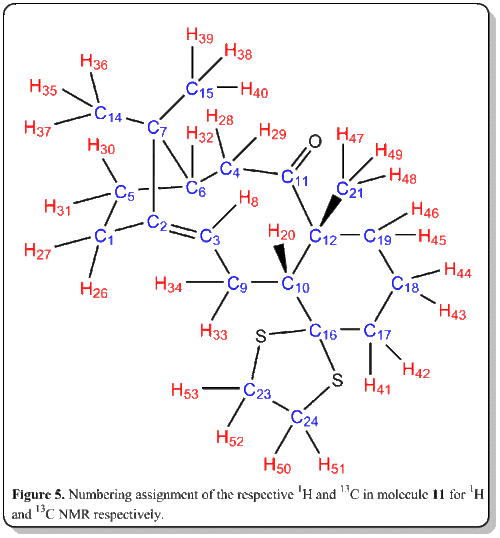
The most useful and rapid technique by far to elucidate the chemical structures would inevitably be Nuclear Magnetic Resonance (NMR).[8] Thus, now we would place computational chemistry in simulating NMR to a test. The 1H and 13C NMR spectrum of molecule 11 was simulated DOI:10042/26522 and compared with literature values[7]. The objective of this comparison was to verify how well the simulated spectra compared to the 'real' spectra, as well as to compare with literature[7] and check on whether the literature values has been correctly interpreted and assigned.
Computational analysis of 11 was executed with the Avogadro program. They were optimized by means of the MMFF94(s) force field option. The alogrithm used was the conjugate gradient option as this evaluates the value of the diagonal terms only while setting all the others to zero. Though it is slower than the steepest descent option, but it assuages the vacillation when reaches less 'steep' gradient where in the steepest descent option, it is very fast but suffer from the vacillation upon reaching less 'steep' gradient. The results and conformation of the computed values are summarized below:
| Types of Total Energy | Molecule 11/ kcal mol-1 | |||
| TOTAL BOND STRETCHING ENERGY | 16.14072 | |||
| TOTAL ANGLE BENDING ENERGY | 44.81234 | |||
| TOTAL STRETCH BENDING ENERGY | 0.76034 | |||
| TOTAL TORSIONAL ENERGY | 12.24163 | |||
| TOTAL OUT-OF-PLANE BENDING ENERGY | 1.05562 | |||
| TOTAL VAN DER WAALS ENERGY | 59.62994 | |||
| TOTAL ELECTROSTATIC ENERGY | -7.80810 | |||
| TOTAL ENERGY | 126.83249 | |||
| Model |
|
Later, the 1H and 13C NMR spectra were simulated with the following procedure. Gaussian was employed to calculate the geometry at the density functional level (DFT) from Avogadro. The calculation was done with the command of Geometry optimization, using B3LYP theory with 6-31G(d,p) basis set. The solvation model was using SCRF(CPCM, Solvent=chloroform).
The results of the computed NMR spectrum is as shown below:
1H NMR
Simulated 1H NMR of molecule 11

Simulated 1H NMR of molecule 11(expanded from 0 - 3.5 ppm)

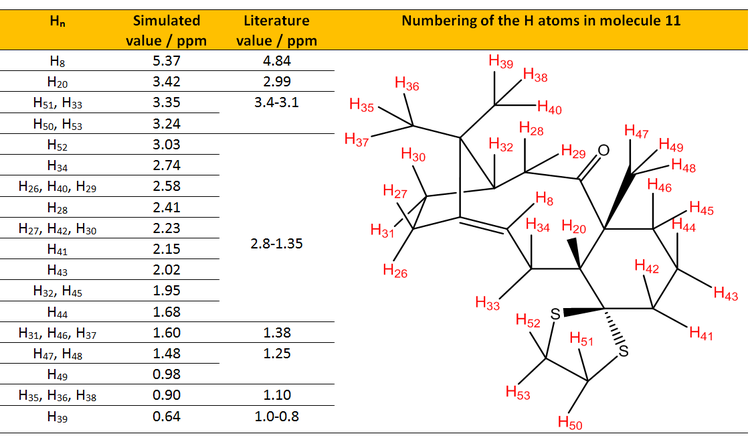
Table 1. Table shows the comparison of assigned simulated 1H NMR values with the literature values.[7]
From Table 1, all of the simulated 1H NMR values were higher than of the reported in experimental value. This could be attributed to the basis set being employed for the calculation which is quite a fundamental (or coarse) basis set and thus lead to the fundamental deviation of the NMR being simulated. The basis set being employed in current calculation was B3LYP/6-31(d,p). As most basis set of quantum chemistry were well-developed for valence electron, NMR on the other hand deals with the electron density closer to the nuclei where causes the current calculation having such error. [9] [10] Also, Malkin [11] [12] had also given explanation on the deviation which will be discussed in the 13C NMR section.
Analysis of the chemical shifts of simulated NMR. From the literature[7], Paquette could only observe a 'bunch' of multiplet in the region from 2.8-1.35 ppm while from the simulated spectrum, we are able to assign it to the individual peaks to individual hydrogens. Again we could observe that the range of the simulated NMR for the protons within the region were all slightly higher than of the literature. The range of simulated NMR was 1.35 ppm while for the literature was 1.45 ppm. From the consideration of the range, the simulated spectrum correlated quite well with the literature. H8 and H20 are protons that are worth analyzing. Both of the values deviated by 0.53 ppm and 0.43 ppm respectively from the literature. This again is attributed to the protons being close to heavy atoms such as sulfur which was not taken into account from the calculation for correction and leading to the deviation. H8 is also highly deshielded (about 5 ppm due to it being an alkene proton) relative to other protons, which is a sensible result and assignment from the both simulation and literature. Other than that, it could be seen that chemical shifts of terminal CH3 protons are not all identical in simulation while in experimental results, they are all identical. This was because in real life NMR experiments, the three protons are rotating about the C-C bond and thus making them to be present in similar chemical environment, consequant to one chemical shift observed only. However, in our simulation, we are able to specifically 'label' and 'fix' the protons at certain coordinates and thus making them not in the same chemical environment. As such, we could see from H47, H48 and H49 having different chemical shifts (1.48ppm and 0.98 ppm) in simulation; while in literature, we could obtain single chemical shift which is 1.25 ppm.
Spin-spin couplings of the 1H NMR
Also, the spin-spin couplings of the 1H NMR was computed as well. Although advances in electronic structure theory (i.e. equation-of-motion coupled cluster theory, second-order polarization propagator approximations) would enable us to predict spin-spin coupling constants with excellent agreement with experimental values, the calculation or computational costs for these methods would be quite costy when it comes to large systems.[13] Thus, we would employ the density functional theory (DFT) method as it is less expensive and relatively quite accurate. [14] For the current study, we have employed the following keywords to execute the calculation:
Basis set: B3LYP/6-311+G(d,p)
Solvent : SCRF(CPCM,solvent=chloroform)
Job: NMR(spinspin, mixed).
The results of the calculation is shown in the DOI link below:
DOI:10042/26640
| nJX-Y | Coupling constant (Simulation), nJX-Y / Hz | Coupling constant (Literature), nJX-Y / Hz | Percentage difference from literature. / % |
| 3JH8-H33 | 11.0 | 7.2 | 53% |
| 3JH8-H34 | 8.8 | 4.7 | 87% |
| 3JH20-H33 | 7.4 | 5.2 | 42% |
| 3JH20-H34 | 11.7 | 6.8 | 72% |
From Table 2, we observe that the spin-spin coupling constant has an extremely massive percentage error from the literature value, up to 87%! This could be attributed to the effect of the spin-orbit effect due to heavy atoms that is not taken into account in the current calculations as our current basis set is not sufficiently to produce a reasonable value and corrections were not being made with respect to the effect. Deng [8] also suggested some modification for calculation of NMR spin-spin coupling constant, where we could employ in our calculations in the future. However, as we proceed to smaller systems such as styrene oxide and methyl-strene oxide in following discussions, we could observe that the simulated values matches quite well with the literature.
13C NMR
Simulated 13C NMR of molecule 11
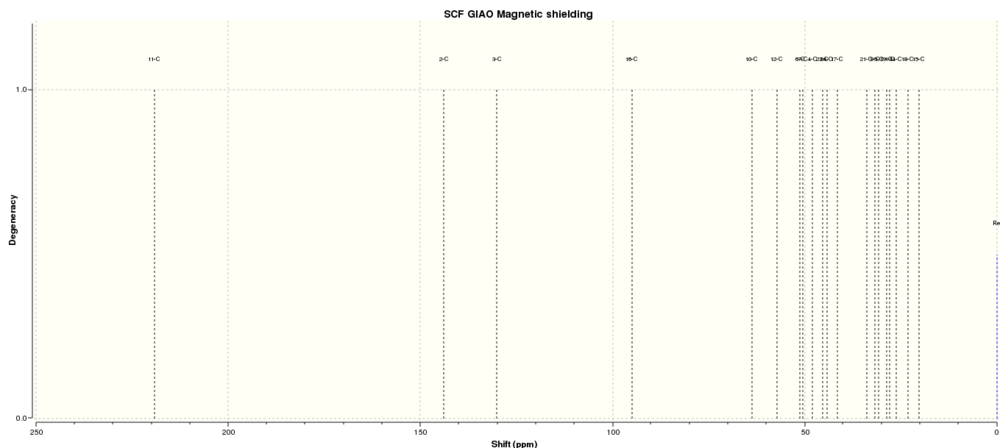
Simulated 13C NMR of molecule 11(expanded from 20 - 250 ppm)

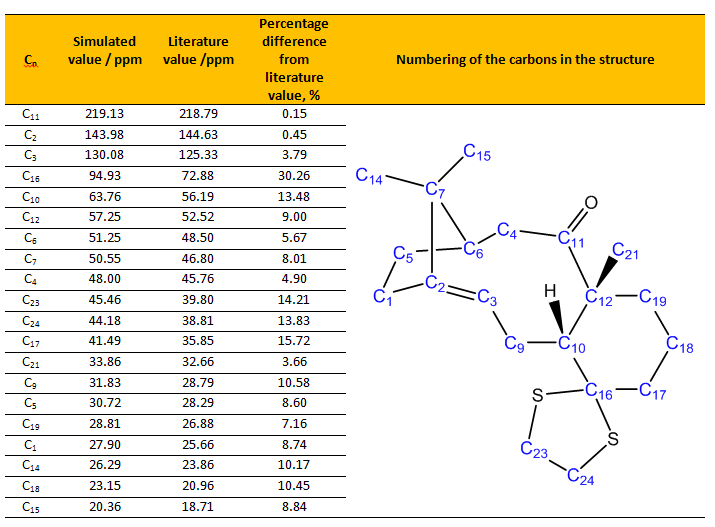
Table 3. Table shows the comparison of assigned simulated 13C NMR values with the literature values.[7]
From Table 3, most of the simulated 13C NMR values have around 0-10% percentage difference from the literature value, where the simulated values are always slightly higher than literature or experimental values. This could be attributed to the factor as discussed before, where the NMR probes more to the core electron density, we instead deal with valence electrons for quantum chemistry basis set. Besides that, Malkin [11] [12] had studied extensively on the simulated NMR values and found that we should also take into account of relativistic effects for transition metal complexes and any systems containing heavy elements (i.e. Cl, I, S and etc) when doing our calculations. The relativistic effects could be categorized into two types, namely scalar and spin-orbit (SO) effects. As NMR parameters depend strongly on the electronic structure near the nucleus of interest, the NMR parameter would be affected by the relativistic effects much rapidly than other properties such as molecular geometries. Thus Malkin [11] [12] [15]had suggested solutions for to correct for the relativistic effects, particularly SO effect which is quite prominent in current study.
There were some simulated values that have more than 10% percentage difference, and for C16, the difference was up to 30% higher than literature value. Also, C10, C17, C23, and C24 values have more than 10% difference than literature value. This could be attributed to the carbons being attached to "heavy" elements, which in this case sulfur, as described previously by Markin[11] [12] [15]. These carbon bearing sulfur have shifts which needed correction for the spin-orbit coupling errors. C16 has much more deviation than C10, C17, C23, and C24. This is glaringly obvious of the reason behind as this carbon has two sulfur atoms attached to it and thus the spin-orbit coupling error would be much more severe. It is noted that C2, C3 and C11 have very high chemical shifts. This is sensible as C2 and C3 are alkene carbons while C11 is next to an oxygen atom, thus they have high chemical shifts and most deshielded. Both the simulation and literature agreed well in terms of assigning for these 13C NMR shifts.
Comparison of relative energies of two isomeric configurations of 11 and its isomer.
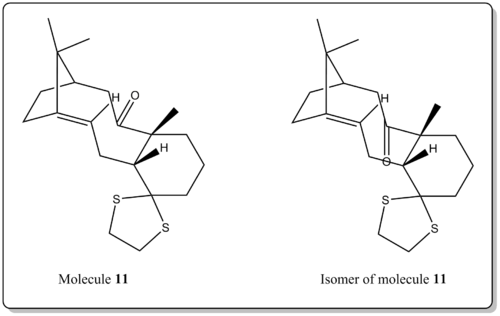
Besides that the free energy, ΔG (Sum of electronic and thermal Free Energies), was computed to compare the relative energies of two isomeric configurations of the molecule 11. (i.e. carbonyl group pointing up or down.)
The results are as shown below:
1) -1651.445143 a.u. (carbonyl group pointing up, as drawn for molecule 11)
Log file: File:Pointingup.log
2) -1651.463260 a.u. (carbonyl group pointing down)
Log file: File:Pointingdown.log
The result is not surprising where when the carbonyl group is pointing down, the molecule is lower in energy, similar to the case of 9 and 10. As explained earlier, it was due to the difference in ring strain imposed on the structure when the carbonyl group is pointing up or down. Also, as there is a thioacetal on the cyclohexane, it locks the cyclohexane into chair form and thus locking the geometry of the larger ring adjacent to it. From our results, it is better when the carbonyl is pointing down as it introduces less strain to the larger the ring due to the constrains in geometry imposed by the adjacent groups of rings.
Part 2
Selected alkenes: styrene and β-methyl styrene.
The crystal structures of Jacobsen's catalyst and Shi's catalyst
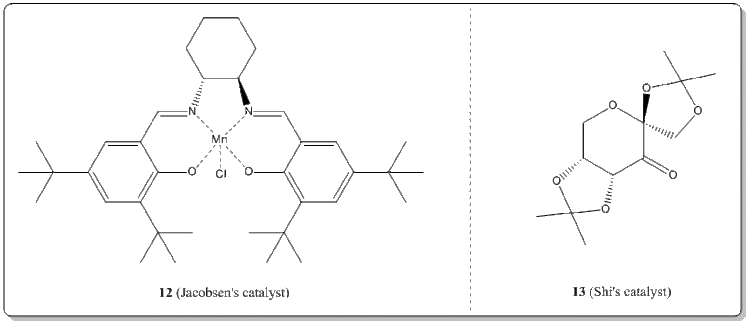
With the Conquest program, the crystal structures of 12 and 13 were searched from the Cambridge Crystal Database (CCDC).
(i) Jacobsen's catalyst, 12. [16]

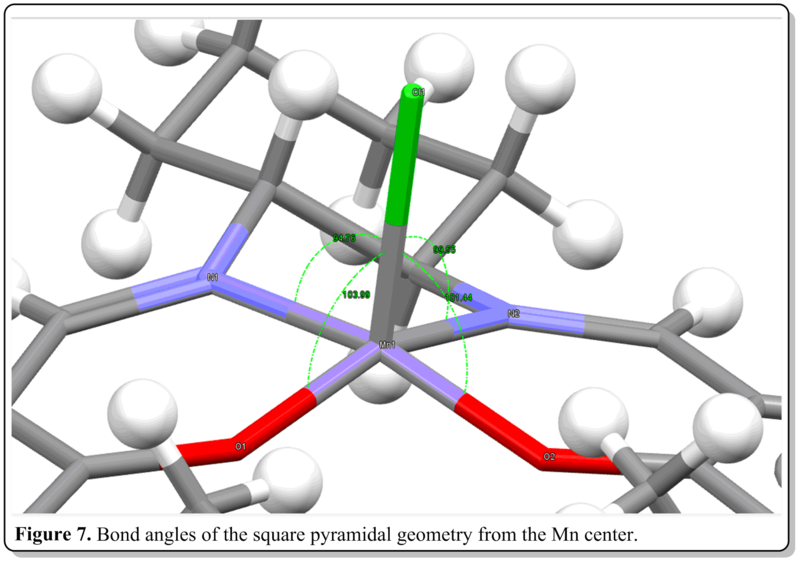
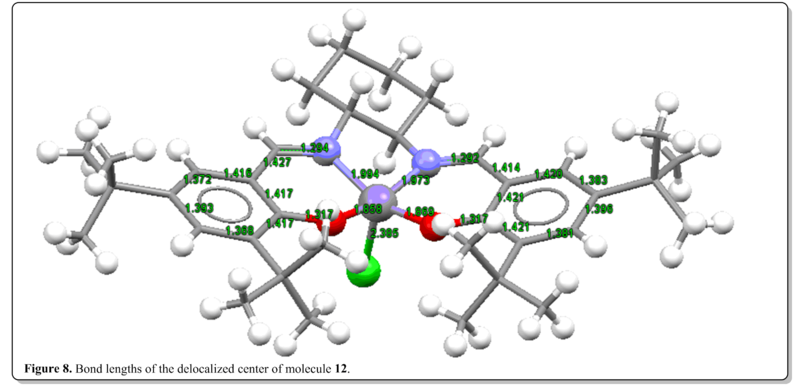
(ii) Shi's catalyst, 13. [17]

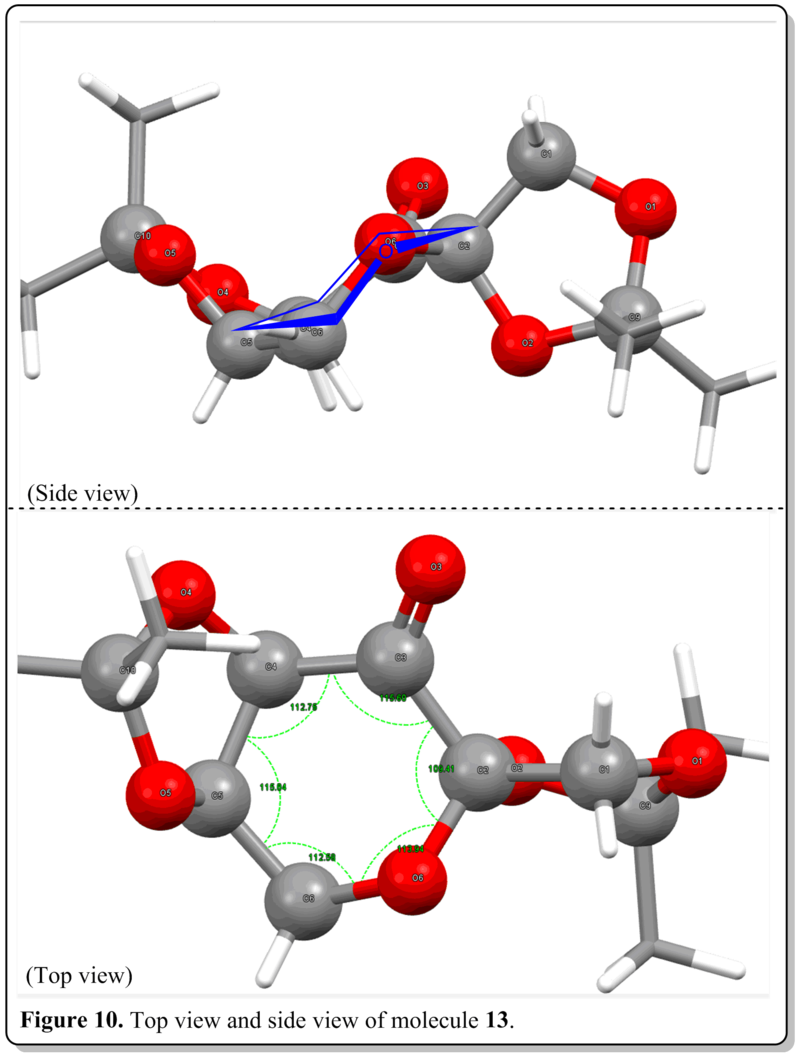
Assigning the absolute configuration of the product
(i) Optical Rotation (OR)
Optical rotation (OR) is one of simplest and easiest methods to characterize a certain chiral molecule. Usually there is no correlation for the quantitative value of OR with certain configuration either R or S of a compound. With the advancement in computational chemistry, we would be able to elucidate or at least have an alternative method to be able to predict the OR of a molecule and from then, hopefully study on the factors determining the OR of chiral molecules.
For our current study, we are using density functional theory (DFT) in the OR prediction. Usage of DFT allows us to perform the calculation at a lower cost compared to other methods and having results with accuracy comparable to correlated wave function methods. In terms of basis set selection, Autschbach [18]also stated that the quality of the basis set is utmost crucial in OR calculation and he suggested that it is vital to incorporate diffuse functions in the basis set. This would allow us to obtain results with a relative median error of c.a. 30%. Thus the OR calculations was executed following from the output of the NMR calculation job. The OR calculation was performed with teh following command:
Basis set: CAM-B3LYP/6-311++g(2df,p) Polarity: polar(optrot) Solvent model:scrf(cpcm,solvent=chloroform) Job type: CPHF=RdFreq <br> Frequencies: 589nm 365nm
(a)(R)-(-)-Styrene oxide, 14 and (R)-(-)-Styrene oxide, 15
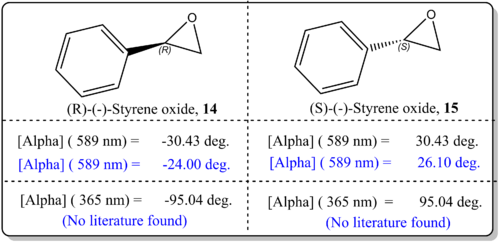
From Figure 11, it was shown that the simulated values agreed well with the literature value. There was just difference by about 6 deg. which indicated that the results are quite reliable. This could be attributed to the basis set being employed for current study CAM-B3LYP/6-311++g(2df,p) which is quite sophisticated. However, Hedegard [21]reported that the best basis set for use with optical rotations is aug-pcS-1, together with the CAM-B3LYP functional which could improve in the calculations. We should also note that the slight deviation from the reported experimental values for OR could be due to the phase model being used in the simulation and experimental values. In the real experiemnts, the OR is taken from the condensed phase, while our computational results refer to gas phase conditions. As such, Kumata[22] had exemplified had shown that the effect of solvation had become severe.
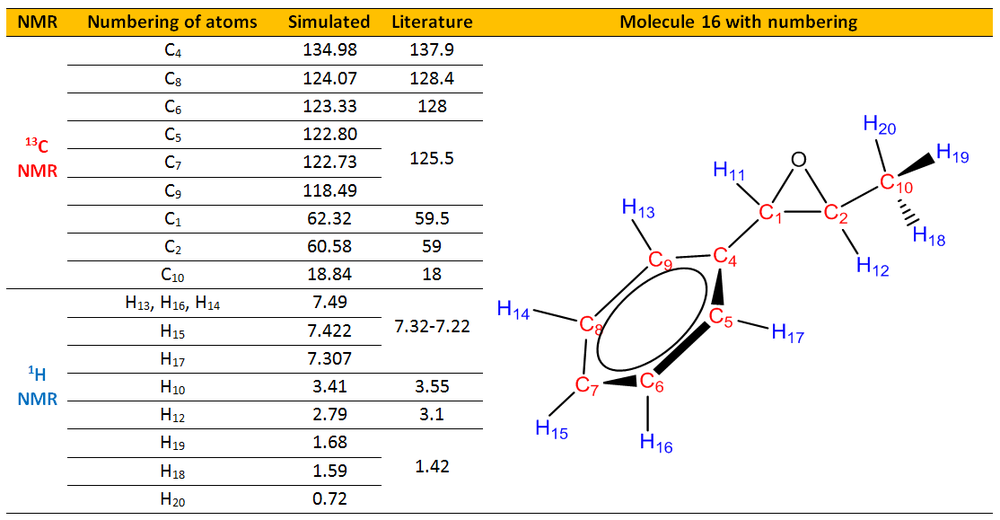
(b)(1R,2R)-(-)-1-Phenylpropylene oxide, 16 and (1S,2S)-(-)-1-Phenylpropylene oxide, 17
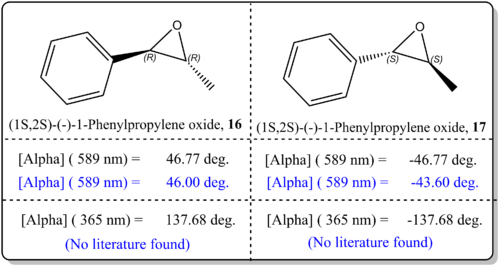
From Figure 12, it was shown that the simulated values agreed well with the literature value. There was just difference by about 4 deg. to 0 deg. which indicated that the results are very reliable. This, similar to the previous case could be attributed to the basis set being employed for current study, CAM-B3LYP/6-311++g(2df,p), which is quite sophisticated. We could enhance on the results by using the method as proposed by Hedegard [21]where he employed the basis set aug-pcS-1, together with the CAM-B3LYP functional for OR calculation. We should also note that the slight deviation from the reported experimental values for OR could be due to the phase model being used in the simulation and experimental values. In the real experiemnts, the OR is taken from the condensed phase, while our computational results refer to gas phase conditions. As such, Kumata[22] had exemplified had shown that the effect of solvation had become severe.
(ii) Vibrational circular dichrosim (VCD)
The calculated NMR properties of compound 14 and 16.
(a) NMR of compound 14.
1H NMR of 14

13C NMR of 14
The simulated 1H and 13C NMR were compared with the literature values.[25]
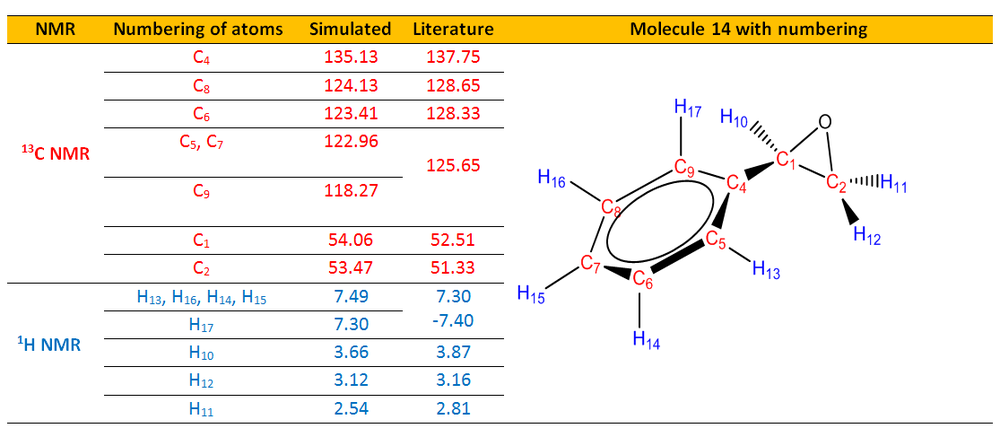
From Table 4, it could be seen that the simulated 13C and 1H NMR chemical shift values matched very well with the 13C and 1H NMR chemical shift experimental values.
Analysis of 13C NMR From both the simulation and literature, C4 to C9 which are carbons in the aromatic ring were assigned with the highest chemical shifts. Meanwhile, C1 has higher chemical shift than C2. These chemical shifts are reasonable and can be rationalized from the fact that C4 to C9 are carbon atoms in the aromatic rings and due to the delocalization of the electron density around the ring,
| nJX-Y | Coupling constant (Simulation), nJX-Y / Hz | Coupling constant (Literature[25]), nJX-Y / Hz | Percentage difference from literature. / % |
| 2JH11-H12 | 5.6 | 5.4 | 4% |
| 3JH12-H10 | 4.4 | 4.0 | 10% |
| 3JH11-H10 | 1.9 | 2.5 | 21% |
(b) NMR of compound 16.
1H NMR of 16
13C NMR of 16
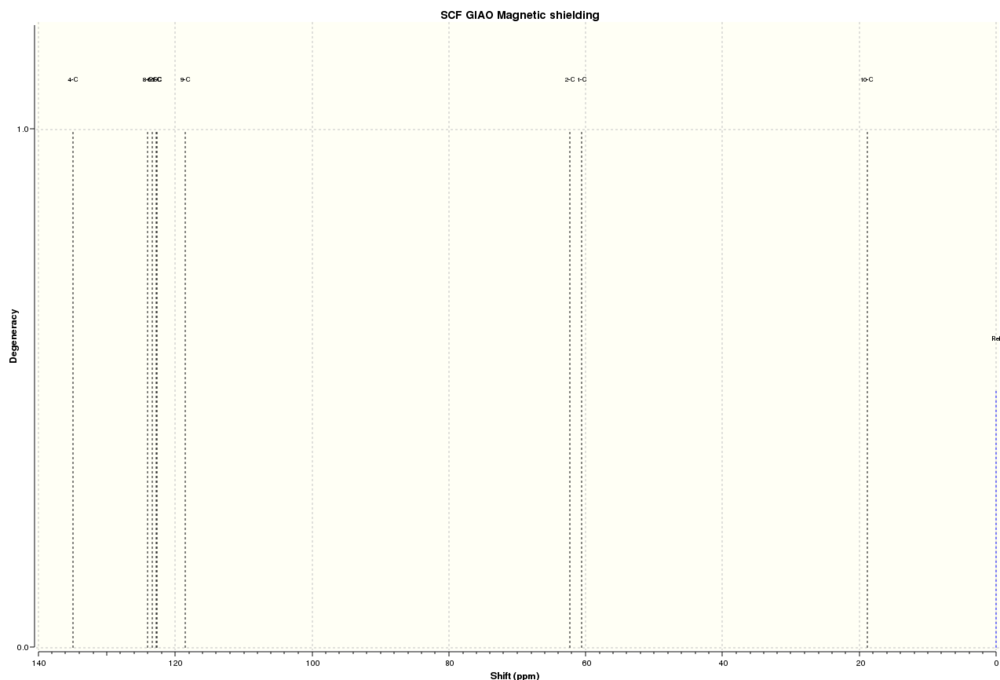
The simulated 1H and 13C NMR were compared with the literature values.[26]
| nJX-Y | Coupling constant (Simulation), nJX-Y / Hz | Coupling constant (Literature[26][27]), nJX-Y / Hz | Percentage difference from literature. / % |
| 3JH11-H12 | 1.4 | 2.5 | 44% |
| 3JH20-H12 | 11.8 | 5.4 | 119% |
| 3JH19-H12 | 3.0 | 5.4 | 44% |
| 3JH18-H12 | 1.4 | 5.4 | 74% |
| 3JH(Average 18, 19, 20)-H12 | 5.4 | 5.4 | 0% |
Using the (calculated) properties of transition state for the epoxidation using Jacobsen's catalyst and Shi's catalyst to determine enantiomeric excess (e.e.)
Using the “sum of electronic and thermal free energies” from the calculations, which corresponds to the Gibbs Free Energy at 298.15K and 1 atm, the transition state for the expoxidation using two of the catalysts are elucidated quantitatively.
(i) Shi's catalyst
| Energies for R,R series, ERR /Hartrees | Energies for S,S Series , ESS/Hartrees |
|---|---|
| -1343.022970 | -1343.017942 |
| -1343.019233 | -1343.015603 |
| -1343.029272 | -1343.023766 |
| -1343.032443 | -1343.024742 |
| Minimum Energy for R,R series, Emin-RR /Hartrees | Minimum Energies for S,S Series, Emin-SS/Hartrees | Difference in the minimum energies, D = Emin-RR- Emin-SS/ Hartrees | Difference in the minimum energies, D = Emin-RR- Emin-SS/ Joules | The ratio of concentrations of the two species, K=exp(-D/RT) | Enantiomeric Excess, ee |
|---|---|---|---|---|---|
| -1343.032443 | -1343.024742 | -0.007701 | -20219 | 3500 | 99.942% |
| Energies for R series, ER /Hartrees | Energies for S Series , ES/Hartrees |
|---|---|
| -1303.730703 | -1303.733828 |
| -1303.730238 | -1303.724178 |
| -1303.736813 | -1303.727673 |
| -1303.738044 | -1303.738503 |
| Minimum Energy for R series, Emin-R /Hartrees | Minimum Energy for S series, Emin-S /Hartrees | Difference in the minimum energies, D=Emin-R- Emin-S/ Hartrees | Difference in the minimum energies, D = Emin-R- Emin-S/ Joules | The ratio of concentrations of the two species, K=exp(-D/RT) | Enantiomeric Excess, ee |
|---|---|---|---|---|---|
| -1303.738044 | -1303.738503 | 0.000459 | 1205 | 0.6148 | -23.851% |
(ii)Jacobsen's catalyst
| Energies for R,R series, ERR /Hartrees | Energies for S,S Series , ESS/Hartrees |
|---|---|
| -3383.253816 | -3383.262481 |
| -3383.254344 | -3383.257847 |
| Minimum Energy for R,R series, Emin-RR /Hartrees | Minimum Energies for S,S Series, Emin-SS/Hartrees | Difference in the minimum energies, D = Emin-RR- Emin-SS/ Hartrees | Difference in the minimum energies, D = Emin-RR- Emin-SS/ Joules | The ratio of concentrations of the two species, K=exp(-D/RT) | Enantiomeric Excess, ee |
|---|---|---|---|---|---|
| -3383.254344 | -3383.262481 | 0.008137 | 21363 | 0.00018 | -99.964% |
| Energies for R series, ER /Hartrees | Energies for S Series , ES/Hartrees |
|---|---|
| -3343.960889 | -3343.969197 |
| -3343.962162 | -3343.963191 |
| Minimum Energy for R series, Emin-R /Hartrees | Minimum Energy for S series, Emin-S/Hartrees | Difference in the minimum energies, D=Emin-R- Emin-S/ Hartrees | Difference in the minimum energies, D = Emin-R- Emin-S/ Joules | The ratio of concentrations of the two species, K=exp(-D/RT) | Enantiomeric Excess, ee |
|---|---|---|---|---|---|
| -3343.962162 | -3343.969197 | 0.007035 | 18470 | 0.000579 | -99.884% |
| Energies for R,S series, ERS /Hartrees | Energies for S,R Series , ESR/Hartrees |
|---|---|
| -3383.25106 | -3383.259559 |
| -3383.25027 | -3383.253443 |
| Minimum Energy for R,S series, Emin-RR /Hartrees | Minimum Energies for S,R Series, Emin-SS/Hartrees | Difference in the minimum energies, D = Emin-RS- Emin-SR/ Hartrees | Difference in the minimum energies, D = Emin-RS- Emin-SR/ Joules | The ratio of concentrations of the two species, K=exp(-D/RT) | Enantiomeric Excess, ee |
|---|---|---|---|---|---|
| -3383.25106 | -3383.259559 | 0.008499 | 22314 | 0.000123 | -99.976% |

Investigating the non-covalent interactions in the active-site of the reaction transition state of Shi's catalyst with beta-methylstyrene
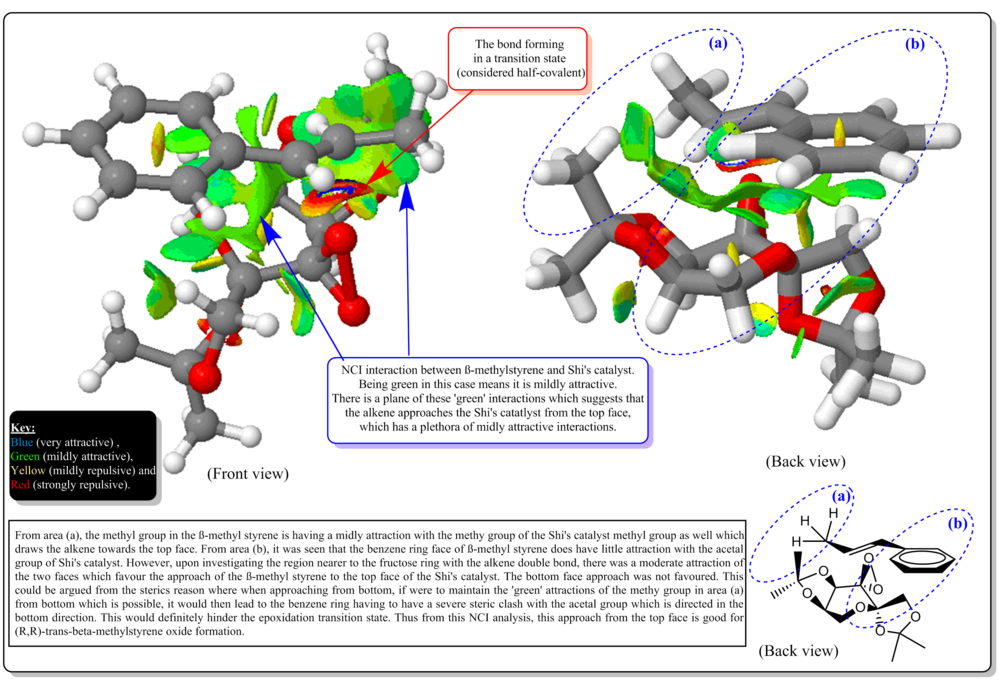
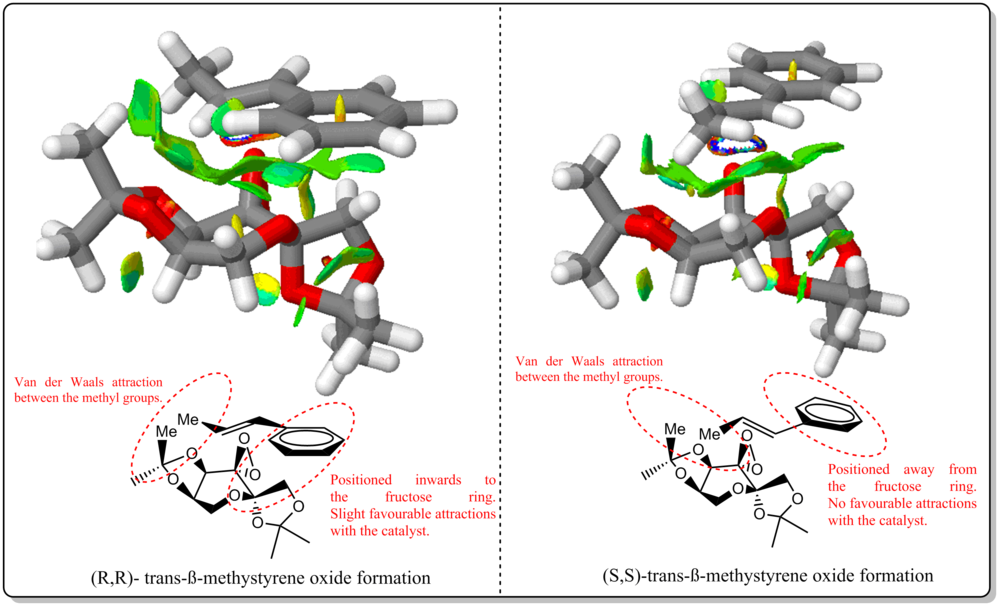
Investigating the Electronic topology (QTAIM) in the active-site of the reaction transition site

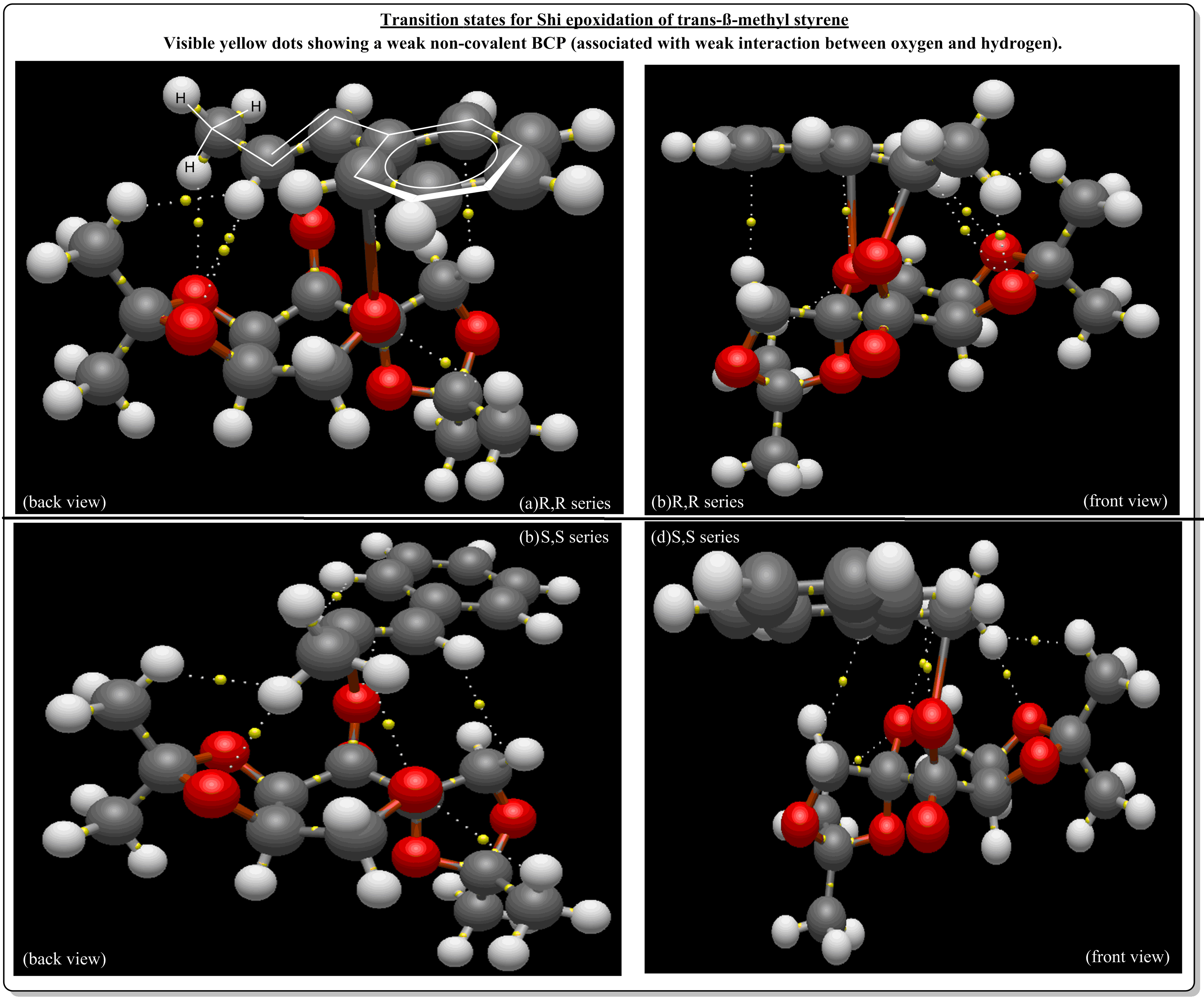
References
- ↑ Wagner, E.C., Hunt, W.C., J. Chem. Edu., 1951, 309-311. DOI:10.1021/ed028p309 10.1021/ed028p309
- ↑ 2.0 2.1 Cristol, S.J., Seifert, W.K., Soloway, S.B., J. Am. Chem. Soc., 1960, 82, 2351.DOI:10.1021/ja01494a060
- ↑ 3.0 3.1 3.2 Alder, K., Stein, G., Justus Liebigs Annalen der Chemie, 1934, 514(1), 1. DOI:10.1002/jlac.19345140102
- ↑ 4.0 4.1 Alonso, F., Yus, M., Tetrahedron Letters, 1996, 37(38), 6925-6928 DOI:10.1016/0040-4039(96)01518-3 Cite error: Invalid
<ref>tag; name "ja2100" defined multiple times with different content - ↑ 5.0 5.1 5.2 Elmore, S.W., Paquette, L.A., Tetrahedron Letters, 1991, 32(3), 319-322. DOI:10.1016/S0040-4039(00)92617-0
- ↑ Bredt, J., Thouet, H., Schmitz, J. Liebigs Ann. Chem., 1924, 437, 1.DOI:10.1002/jlac.19244370102
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Paquette,L.A., Pegg, N.A., Toops, D., Maynard, G.D., Rogers, R.D., J. Am. Chem. Soc., 1990, 112(1), 277-283. DOI:10.1021/ja00157a043 Cite error: Invalid
<ref>tag; name "ja2093" defined multiple times with different content - ↑ 8.0 8.1 Deng, W., J.Chem.Theory Comput., 2006, 2, 1028-1037. DOI:10.1021/ct600110u
- ↑ Helgaker, T., Jazunski, M., Ruud, K., Gorska, A., Theor. Chem, Acc., 1998, 99(3), 175-182.DOI:10.1007/s002140050321
- ↑ Oddershede, J., Geertsen, J., Scuseria, G.E., J. Phys. Chem.,1988,92,3056-3059. DOI:10.1021/j100322a009
- ↑ 11.0 11.1 11.2 11.3 Malkin, V.G., Malkina, O.L., Salahub, Chem. Phys. Letters, 1996, 261, 335-345. DOI:10.1016/0009-2614(96)00988-8
- ↑ 12.0 12.1 12.2 12.3 Malkina, O.L., Schimmelpfenning, B., Kaupp, M., Hess, B.A., Chandra, P., Wahlgren, U., Malkin, V.G., Chem. Phys. Letters, , 1998, 296, 93. DOI:10.1016/S0009-2614(98)00998-1
- ↑ Cheeseman, J.R., Frisch, M.J., J.Chem.Theory Comput., 2006, 2, 1028-1037. DOI:10.1021/ct600110u
- ↑ Helgaker, T., Watson, M., Handy, N. C., J. Chem. Phys., 2000, 113, 9402-9409. DOI:10.1063/1.1321296
- ↑ 15.0 15.1 Malkin, V.G., Malkina, O.L., Kaupp, M., Chem. Phys. Letters, 1997, 265, 55-59. DOI:10.1016/S0009-2614(96)01425-X
- ↑ Yoon,J.W., Yoon,T.-S., Lee, S.W., Shin, W., Acta Crystallogr.,Sect.C:Cryst.Struct.Commun., 1999, 55, 1766. DOI:10.1107/S0108270199009397
- ↑ Wang, Z.-X., Miller, S.M., Anderson, O.P., Shi, Y., J.Org.Chem., 2001, 66, 521. DOI:10.1021/jo001343i
- ↑ Autschbach, J., Patchkovskii, S., Ziegler, T., Gisbergen, S.G.V., Baerends, E. J., J. Chem. Phys., 2002, 117, 581.DOI:10.1063/1.1477925
- ↑ Capriati, V., Florio, S., Luisi, R., Salomone, A., Org. Lett., 2002, 4 (14), 2445–2448. DOI:10.1021/ol026212d
- ↑ Matsumoto, K., Kubo, T. and Katsuki, T., Chem. Eur. J., 2009, 15, 6573–6575.DOI:10.1002/chem.200901048
- ↑ 21.0 21.1 Hedegård, Jensen, F., Kongsted, J., J. Chem. Theory Comput., 2012, 8 (11), 4425–4433. DOI:10.1021/ct300359s
- ↑ 22.0 22.1 Kumata, Y.; Furukawa, J.; Fueno, T. Bull. Chem. Soc. Jpn. 1970,43, 3920−3921. DOI:10.1246/bcsj.43.3920
- ↑ Besse, P., Benabd, M.F., et. al., Tetrahedron: Asymmetry, 1994, 5(7), 1249-1268.DOI:10.1016/0957-4166(94)80167-3
- ↑ Koya, S., Nishioka, Y., Mizoguchi, H., Uchida, T. and Katsuki, T., Angew. Chem. Int. Ed., 2012, 51, 8243–8246.DOI:10.1002/anie.201201848
- ↑ 25.0 25.1 25.2 Solodenkoa, W., Jasb, G., Kunzc, U., Kirschning, A., Synthesis, 2007, 4, 583-589. DOI:10.1055/s-2007-965877
- ↑ 26.0 26.1 Jia, L., Wanga, Y.-N., Qiana, C., Chena, X.-Z., Syn. Comm., 2013, 43(16), 2256-2264. DOI:10.1080/00397911.2012.699578
- ↑ Page,P.C.B., Parker, P., Buckley, B.R., Rassias, G.A., Bethell, D., Tetrahedron, 2009, 65, 2910–2915. DOI:10.1016/j.tet.2009.02.007
