Rep:Mod:yqc01506162
NH3 molecule
Summary information
| Calculation method | RB3LYP |
|---|---|
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) in au | -56.55776873 |
| RMS gradient norm in au | 0.00000485 |
| Point group | C3V |
Item section
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Jmol of NH3
test molecule |
The optimisation file is liked to File:YQC18 NH3 OPTF POP.LOG
Key structural information
N-H bond distance= 1.02 +/- 0.01Å
H-N-H bond angle= 106 +/- 1°
Display vibrations table
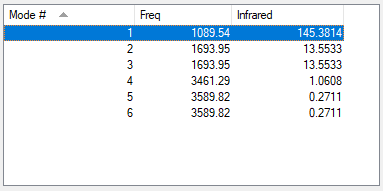
Table of vibration and intensities
| wavenumber(cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| intensity(arbitary units) | 145 | 13.6 | 13.6 | 1.06 | 0.271 | 0.271 |
| image |  |
 |
 |
 |
 |
|
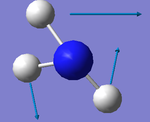
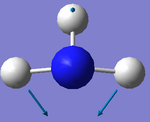
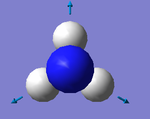
1. 6 modes are expected from 3N-6 rule
2. Modes with wavenumbers both being 1694 cm-1 are degenerate. Modes with wavenumbers both being 3590 cm-1 are also degenerate.
3. Modes with wavenumber 1090, 1694 and 1694 cm-1 are bending vibrations. Modes with wavenumbers 3461, 3590 and 3590 cm-1 are "bond stretch" vibrations.
4. Mode with wavenumber 3461 cm-1 is highly symmetric.
5. Mode with wavenumber 1090 cm-1 is the "umbrella mode"
6. I would expect to see 2 bands in an experimental spectrum of gaseoous ammonia.
NBO charges
The charge on N atom is -1.125 and charge on H atom is 0.375. I would expect negative charge on N and positive charge on H. This is because N is more electronegative than H hence pulls electron density towards H.
N2 and H2 molecules
N2 molecule
Summary information
| Calculation method | RB3LYP |
|---|---|
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) in au | -109.52412868 |
| RMS gradient norm in au | 0.00000060 |
| Point group | D∞h |
Item section
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Jmol of N2
test molecule |
The optimisation file is liked to File:YQC N2 OPTF POP.LOG
Key structural information
Optimised N-N bond length= 1.11 +/- 0.01 Å
Optimised N-N bond angle= 180 +/- 1°
Display vibration table
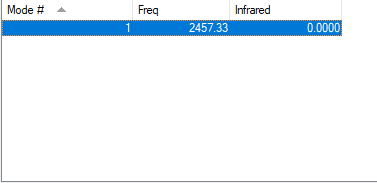
Table of vibrations and intensities
| wavenumber(cm-1) | 2457 |
|---|---|
| symmetry | SGG |
| intensity(arbitary units) | 0.00 |
| image | 
|
NBO charges
The charge on both N atom is 0.000
H2 molecule
Summary information
| Calculation method | RB3LYP |
|---|---|
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) in au | -1.17853936 |
| RMS gradient norm in au | 0.00000017 |
| Point group | D∞h |
Item section
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Jmol of H2
test molecule |
The optimisation file is liked to File:YQC H2 OPTF POP.LOG
Key stuctural information
Optimised H-H bond length = 0.74 +/- 0.01 Å
Optimised H-H bond angle = 180 +/- 1°
Display vibration table
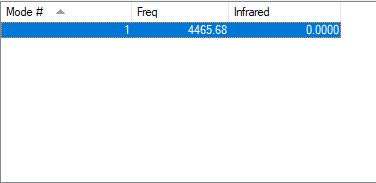
Table of vibration and intensities
| wavenumber(cm-1) | 4466 |
|---|---|
| symmetry | SGG |
| intensity(arbitary units) | 0.00 |
| image | 
|
NBO charges
The charge on both N atom is 0.000.
A mono-metallic TM complex that coordinates N2
| molecule name | (Dinitrogen-N)-(2-(1-(2,6-di-isopropylphenylimino)ethyl)-6-(1-(2,6-di-isopropylphenyl)amidoehenyl)pyridine-N,N'N)-iron |
|---|---|
| unique identifier | COGBIK |
| N-N bond distance obtained(Å) | 1.11 |
| N-N bond distance in the complex(Å) | 1.14 |
| structure | 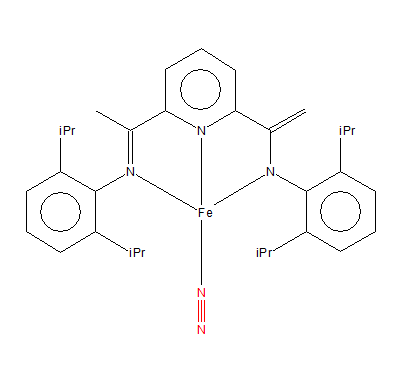
|
| Link to the structure | https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=COGBIK&DatabaseToSearch=Published |
The N-N bond distance in the complex is longer than computational distance.
Experimental reason: After accepting electrons from N, Fe becomes very negative and stabilizes by donating 3d electrons into pi-antibonding orbitals on N atom. This shortens Fe-N bond hence elongates N-N bond. This is called 'synergic effect'.
Computational reason: Computational analysis errors
Haber-Bosch reaction energy calculation
E(NH3)= -56.5577687 a.u
2*E(NH3)= -113.1155375 a.u
E(N2)= -109.5241287 a.u
E(H2)= -1.1785394 a.u
3*E(H2)= -3.5356181 a.u
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.5 kJ/mol
The ammonia product is more stable
My choice of small molecule: HCN
Summary table
| Calculation method | RB3LYP |
|---|---|
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) in au | -93.42458132 |
| RMS gradient norm in au | 0.00017006 |
| Point group | C∞v |
Item section
Item Value Threshold Converged? Maximum Force 0.000370 0.000450 YES RMS Force 0.000255 0.000300 YES Maximum Displacement 0.000676 0.001800 YES RMS Displacement 0.000427 0.001200 YES
Jmol of HCN
test molecule |
The optimisation file is liked to File:YQC HCN OPTF POP.LOG
Key strucutral information
Optimised C-H bond length= 1.07 +/- 0.01 Å
Optimised C-N bond length = 1.16 +/- 0.01 Å
Optimised H-C-N bond angle = 180 +/- 1°
Display vibrations table
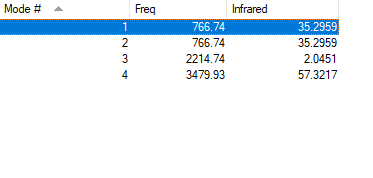
Table of vibrations and intensities
| wavenumber(cm-1) | 2215 | 767 | 767 | 3480 |
|---|---|---|---|---|
| symmetry | SG | PI | PI | SG |
| intensity(arbitary units) | 2.05 | 35.3 | 57.3 | |
| image |  |
 |
 |
|
NBO charges
The charge on N atom is -0.308
The charge on C atom is 0.073
The charge on H atom is 0.234
Molecular orbitals
1.

1s atomic orbital from nitrogen contributes to the MO. The MO is non-bonding. The MO is deep in energy. The MO is occupied and has no effect on bonding.
2.
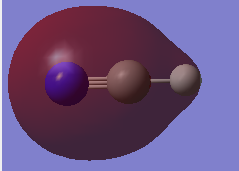
2s atomic orbitals from carbon,nitrogen and 1s atomic orbital from hydrogen contribute to the MO. The MO is bonding. The MO is deep in energy. The MO is occupied and stabilizes the bonding.
3.

2s atomic orbitals from hydrogem and nitrogen and 2pz atomic orbital from carbon contribute to the MO. The MO is bonding. The MO is deep in energy. The MO is occupied and destabilizes the bonding.
4.
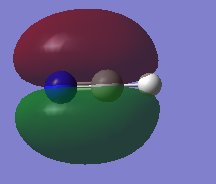
2py atomic orbitals from carbon and nitrogen contribute to the MO. The MO is bonding. The MO is in the LUMO. The MO is occupied and stabilizes the bonding.
5.
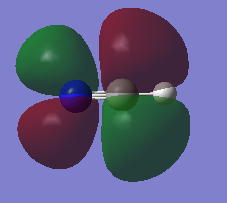
2py atomic orbitals from carbon and nitrogen contribute to the MO. The MO is antibonding. The MO is in the HOMO. The MO is unoccupied and therefore has no effect on bonding.
Independent work: Cl2 molecule
Summary table
| Calculation method | RB3LYP |
|---|---|
| Basis set | 6-31G(d.p) |
| Final energy E(RB3LYP) in au | -920.34987886 |
| RMS gradient norm in au | 0.0000251 |
| Point group | D∞h |
Item section
Item Value Threshold Converged? Maximum Force 0.000043 0.000450 YES RMS Force 0.000043 0.000300 YES Maximum Displacement 0.000121 0.001800 YES RMS Displacement 0.000172 0.001200 YES
Jmol of Cl2
test molecule |
The optimisation file is liked to File:YQC CL2 OPTF POP.LOG
Key structural information
Optimised Cl-Cl bond length= 2.04 +/- 0.01 Å
Optimised Cl-Cl bond angle= 180 +/- 1°
Display vibrations table
Table of vibration and intensities
| Wavenumber(cm-1) | 520 |
|---|---|
| Symmetry | SGG |
| Intensity(arbitary units) | 0.00 |
| image |
NBO charges
The charge on both Cl atom is 0.000.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES, however you have left all the jmol captions as the default “test molecule” this gives the reader no information.
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, However you have given a bond angle of 180 for N2 and H2, there are no bond angles in diatomic molecules. Bond angles involve exactly 3 atoms.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 2.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, you have included the relevant information well done! Your explanations of MOs 1 and 3 are very good and clearly explained! On the charges you could have improved by explaining why each atom has a different charge in terms of the relative electronegativities of the elements. For MO 4 the C-H bond is strengthened but the N-C bond is weakened. You also seem to be confused on the terms HOMO and LUMO, have a look back in your lecture notes for those terms or show your tutor your wiki and ask them.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You did an extra calculation on Cl2 well done!