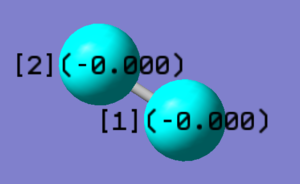Rep:Mod:yh13018
NH3 molecule
Summary information
- H3 molecule
- Calculation Method: RB3LYP
- Basis Set:6-31G(d,p)
- final energy E(RB3LYP) in atomic units (au):-56.55776873
- Point group:C3V
File:HYY PHUNT NH3 OPTF POP.LOG
"Item" section
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986262D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
JMol of structure
test molecule |
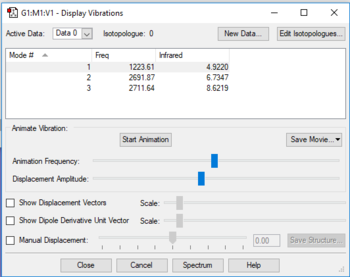
The display vibrations table
The wavenumber, symmetry and intensity of each vibration
| wavenumber cm-1 | 1090 | 1694 | 1694 | ||
|---|---|---|---|---|---|
| symmetry | A1 | E | E | ||
| intensity arbitrary units | 145 | 14 | 14 | ||
| image |  |
 |
 |
| wavenumber cm-1 | 3461 | 3590 | 3590 | ||
|---|---|---|---|---|---|
| symmetry | A1 | E | E | ||
| intensity arbitrary units | 1 | 0.3 | 0.3 | ||
| image |  |
 |
 |
- 6 modes are expected from the 3N-6 rule.
- 2 and 3 are degenerate; 5 and 6 are degenerate.
- 1, 2 and 3 are bending. 4, 5 and 6 are bond stretch.
- 1 and 4 are highly symmetric
- 1 is umbrella mode
- 4 bands are expected to see in an experimental spectrum of gaseous ammonia
Charge distribution
N is negatively charged which is -1.125 and H is positively charged which is 0.375 according to the data. N has higher electronegativity than H thus N attracts electrons from H. In this case, N is more negative and H is more positive
H2 molecules
Summary information
- H2 molecule
- Calculation Method: RB3LYP
- Basis Set:6-31G(d,p)
- final energy E(RB3LYP) in atomic units (au):-1.17853936
- Point group:D*H
"Item" section
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.167770D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
JMol of structure
test molecule |
The display vibrations table
The wavenumber, symmetry and intensity of each vibration
| wavenumber cm-1 | 4466 | ||
|---|---|---|---|
| symmetry | SGG | ||
| intensity arbitrary units | 0 | ||
| image |  |
Charge distribution
N2 molecule
Summary information
- N2 molecule
- Calculation Method: RB3LYP
- Basis Set:6-31G(d,p)
- final energy E(RB3LYP) in atomic units (au):-109.52412868
- Point group: D*H
"Item" section
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.383593D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
JMol of structure
test molecule |
The display vibrations table
The wavenumber, symmetry and intensity of each vibration
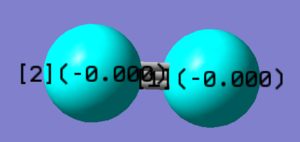
| wavenumber cm-1 | 2457 | ||
|---|---|---|---|
| symmetry | SGG | ||
| intensity arbitrary units | 0 | ||
| image |  |
Charge distribution
A mono-metallic TM complex that coordinates N2
Bis(dinitrogen)-(1,1,4,8,11,11-hexaphenyl-1,4,8,11-tetraphosphaundecane)-molybdenum structure AQUVOY (The link is https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=AQUVOY&DatabaseToSearch=Published) and the two N-N distances are 1.12(1) and 1.11(1) Å. Compared with the bond length of N2 (1.05(5)Å), they are slightly different and the length in the structure found is longer. As one N is bonded to Mo, the electrons on it not only share with other nitrogen but also with Mo thus less number of bonded electrons and lower bond order. Therefore the less stronger the pull and the longer the bond.
the energy for the reaction of N2 + 3H2 -> 2NH3 (Haber-Bosch process)
E(NH3)=-56.55776873 au
2*E(NH3)=-113.1155375 au '
E(N2)=-109.52412868 au
E(H2)=-1.17853936 au
3*E(H2)=-3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579074 au = -146.04785879 kJ/mol
H2S
Summary information
- H2S molecule
- Calculation Method: RB3LYP
- Basis Set:6-31G(d,p)
- final energy E(RB3LYP) in atomic units (au):-399.39162414
- Point group: C2V
"Item" section
Item Value Threshold Converged?
Maximum Force 0.000175 0.000450 YES
RMS Force 0.000145 0.000300 YES
Maximum Displacement 0.000380 0.001800 YES
RMS Displacement 0.000390 0.001200 YES
Predicted change in Energy=-1.212995D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3474 -DE/DX = 0.0002 !
! R2 R(1,3) 1.3474 -DE/DX = 0.0002 !
! A1 A(2,1,3) 92.681 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
JMol of structure
test molecule |
The display vibrations table
The wavenumber, symmetry and intensity of each vibration
| wavenumber cm-1 | 1224 | 2692 | 2712 | ||
|---|---|---|---|---|---|
| symmetry | A1 | A1 | B2 | ||
| intensity arbitrary units | 5 | 7 | 9 | ||
| image |  |
 |
 |
Charge distribution
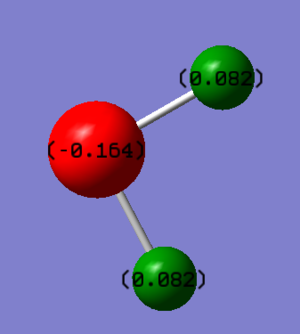
H2S MO
H2S molecule is polar because there are two lone pairs on S. The lone pairs repulse the bonding pairs thus the shape is bent. S is slight more electronegative than H, therefore S is more negatively charged., which is -0.164 and H is more positively charged which is 0.082. The atomic orbitals on sulphur are able to accommodate the 1s orbital from two hydrogens without hybridisation.
The diagram shows the bonding orbital which is from the 2px orbital. It participates in the molecular bonding of H2S and it is occupied. The energy is -183.24569ev
The diagram shows the bonding orbital which is from the 2py orbital. It participates in the molecular bonding of H2S and it is occupied. The energy is -183.18844ev
The diagram shows the bonding orbital which is from the 2pz orbital. It participates in the molecular bonding of H2S and it is occupied. The energy is -183.06884ev
The orbitals are all atomic orbitals above.
The highest occupied molecular orbital in highest energy among all molecular occupied orbitals which is -11.57353ev, is shown above. The orbital is contributed from 3pz orbital which has no bonding with 1s orbital of hydrogen. Both electrons come from sulphur and it results in one lone pair.
The lowest unoccupied molecular orbital in lowest energy among all molecular unoccupied orbitals which is 3.39674ev, is shown above. It is an antibonding orbital which is contributed from 1s orbital of hydrogen and 3py orbital of sulphur. In this case, the probability of finding electrons and the density of electrons are much lower in the orbital. Therefore the energy considerably increases (from -11.57353ev to 3.39674ev), which makes it hard to form a bond.
The graph demonstrates the molecular bonding orbital which is contributed from the 3s orbital of sulphur and 1s orbital of hydrogen. The orbital is occupied and it is in a lower energy level compared with the molecular orbitals below which is -26.82129ev so it is occupied first.
The graph demonstrates the molecular bonding orbital which is contributed from the 3px orbital of sulphur and 1s orbital of hydrogen. The orbital is occupied and it is in a second lower energy level which is -15.19887ev
The graph demonstrates the molecular bonding orbital which is contributed from the 3px orbital of sulphur and 1s orbital of hydrogen. The orbital is occupied and it is in a third lower energy level which is -13.58420ev
According to Aufbau and Paulli exclusion principles, the four molecular bonding orbitals are occupied from the one with the lowest energy to the one with the highest energy. Unlike the LUMO, the probability and density is high thus it is to form bonding.
Extra calculation on another small molecule
CH4 molecule
Summary information
- CH4 molecule
- Calculation Method: RB3LYP
- Basis Set:6-31G(d,p)
- final energy E(RB3LYP) in atomic units (au):-56.55776873
- Point group:TD
"Item" section
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000179 0.001800 YES
RMS Displacement 0.000095 0.001200 YES
Predicted change in Energy=-2.256043D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.092 -DE/DX = -0.0001 !
! R2 R(1,3) 1.092 -DE/DX = -0.0001 !
! R3 R(1,4) 1.092 -DE/DX = -0.0001 !
! R4 R(1,5) 1.092 -DE/DX = -0.0001 !
! A1 A(2,1,3) 109.4712 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4712 -DE/DX = 0.0 !
! A3 A(2,1,5) 109.4712 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.4712 -DE/DX = 0.0 !
! A5 A(3,1,5) 109.4712 -DE/DX = 0.0 !
! A6 A(4,1,5) 109.4712 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -120.0 -DE/DX = 0.0 !
! D2 D(2,1,5,3) 120.0 -DE/DX = 0.0 !
! D3 D(2,1,5,4) -120.0 -DE/DX = 0.0 !
! D4 D(3,1,5,4) 120.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
JMol of structure
test molecule |
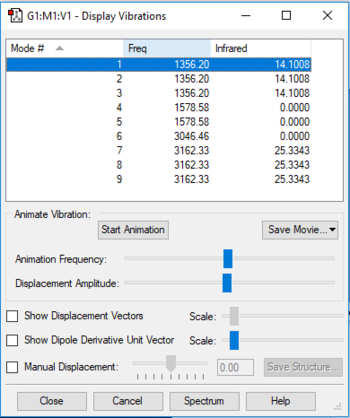
The display vibrations table
The wavenumber, symmetry and intensity vibrations
Charge distribution
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 0/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES - good structure.
Do you effectively use tables, figures and subheadings to communicate your work?
YES, however you have used the caption "test molecule" for all of your jmols which is not a useful caption for the reader.
Also you have used generic file names for your MO image files and therefore one of them is already referring to an image which I don't think you meant to include.
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - good answers for the charges question and most of the vibration questions.
However due to the low intensity of vibrations 4, 5 and 6 you only see two peaks in the IR spectrum.
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES - however you got the wrong bond length for the computed bond. Your results show an N-N bond of 1.105 not 1.05 as you wrote.
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - you explained the charges well, well done.
However you have referred to the 2p AOs on the sulphur atom as "bonding" this is incorrect and these AOs are non-bonding.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
you have completed an extra calculation on ch4, well done.
Do some deeper analysis on your results so far