Rep:Mod:wsk811
Week 1 Inorganic computational
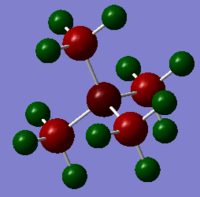
BH3
BH3 optimisation
| File Name | SW_BH3_OPT | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 3-21G | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.46226429 | a.u. |
| RMS Gradient Norm | 0.00008851 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0003 | Debye |
| Point Group | CS | |
| Job cpu time: 0 days 0 hours 0 minutes 20.0 seconds. |
BH3 Optimisation
Item Value Threshold Converged?
Maximum Force 0.000220 0.000450 YES
RMS Force 0.000106 0.000300 YES
Maximum Displacement 0.000940 0.001800 YES
RMS Displacement 0.000447 0.001200 YES
Predicted change in Energy=-1.672479D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1948 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1947 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1944 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0157 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.986 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9983 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
| File Name | SW_bh3_opt_631G | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532358 | a.u. |
| RMS Gradient Norm | 0.00008206 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0003 | Debye |
| Point Group | CS | |
| Job cpu time: 0 days 0 hours 0 minutes 14.0 seconds. | ||
| Optimised Bond Length | 1.19, 1.19, 1.19 | Å |
| Optimised Bond Angle | 120.0, 120.0, 120.0 |
BH3 Optimisation 631G
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000099 0.000300 YES
Maximum Displacement 0.000875 0.001800 YES
RMS Displacement 0.000418 0.001200 YES
Predicted change in Energy=-1.452086D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1928 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1926 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1924 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0146 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9866 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9988 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BH3 optimisation 631G 2nd time
| File Name | BH3_OPT_3 | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532364 | a.u. |
| RMS Gradient Norm | 0.00000000 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 9.0 seconds. | ||
| Optimised Bond Length | 1.19, 1.19, 1.19 | Å |
| Literature Bond Lengthlit 2 | 1.2325 | Å |
| Optimised Bond Angle | 120.0, 120.0, 120.0 |
BH3 Optimisation 2nd time
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000000 0.000006 YES
RMS Displacement 0.000000 0.000004 YES
Predicted change in Energy=-1.214719D-18
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BH3 Frequency
| File Name | SW_BH3_FREQ | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532358 | a.u. |
| RMS Gradient Norm | 0.00008199 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0003 | Debye |
| Point Group | CS | |
| Job cpu time: 0 days 0 hours 0 minutes 11.0 seconds. |
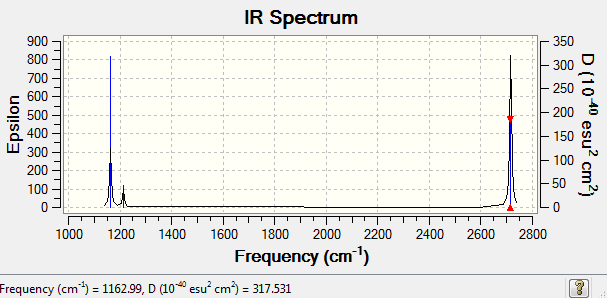
BH3 Frequency Low frequencies --- 0.0006 0.0007 0.0007 33.8245 41.5797 43.6874 Low frequencies --- 1163.5016 1213.4680 1213.5880
comment since the frequency is too high. Therefore point group constraint is added to run a 2nd time.
BH3 Frequency 2nd
| File Name | BH3_FREQ_3 | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532364 | a.u. |
| RMS Gradient Norm | 0.00000001 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 6.0 seconds. |
BH3 Frequency Low frequencies --- -9.3741 -9.3588 -0.0753 0.0007 0.5350 2.4499 Low frequencies --- 1162.9902 1213.1495 1213.1497
BH3 Vibration
In your wiki also explain why are there less than six peaks in the spectrum, when there are obviously six vibrations.
There are indeed six vibrational modes shown in the table. However, the No.2 and No.3 both have frequency at 1213 cm-1 and No. 5 and No. 6 both have the same frequency at 2716 cm-1. So they overlap to each other and only appear two peaks. On the other hand, the vibration of No. 4 is zero, as it is symmetrical stretching and no change in dipole. So it is IR inactive. Therefore there are only 3 peaks appear in the spectrum.
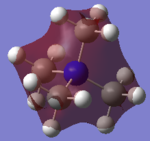
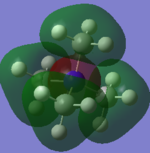
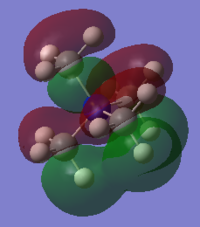
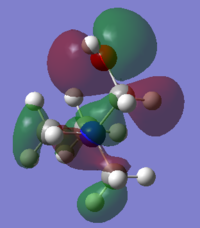
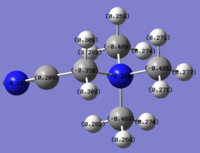
Molecular Orbital of BH3
[Molecular Orbital of BH3 D space]
Answer the following questions:
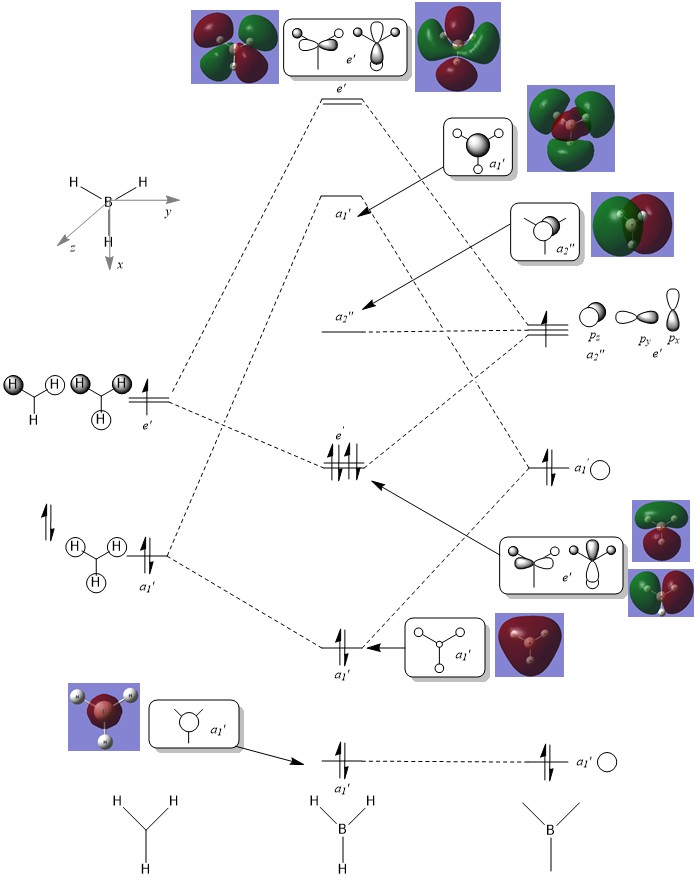
Are there any significant differences between the real and LCAO MOs?
What does this say about the accuracy and usefulness of qualitative MO theory?
As can be seen from the diagram, the calculated MO diagram matches quite well with the MO theory diagram, apart from MO theory shows the atomic orbitals and its interactions while Gaussview merges all the orbitals together. Overall, it is a pretty good assumption. However, the limitation takes place when the molecule becomes very complicated. So it will be very hard to draw the MO diagrams out by LCAO.
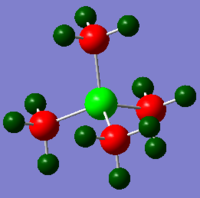
GaBr3
GaBr3 optimisation
| File Name | SW_GABR3_OPT | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -41.70082783 | a.u. |
| RMS Gradient Norm | 0.00000016 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 6.0 seconds. | ||
| Optimised Bond Length | 2.35, 2.35, 2.35 | Å |
| Literature Bond Lengthlit 2 | 2.3525 | Å |
| Optimised Bond Angle | 120.0, 120.0, 120.0 |
GaBr3 optimisation
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
Predicted change in Energy=-1.282691D-12
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.3502 -DE/DX = 0.0 !
! R2 R(1,3) 2.3502 -DE/DX = 0.0 !
! R3 R(1,4) 2.3502 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
GaBr3 Frequency
| File Name | SW_GABR3_FREQ | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -41.70082783 | a.u. |
| RMS Gradient Norm | 0.00000011 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 6.0 seconds. |
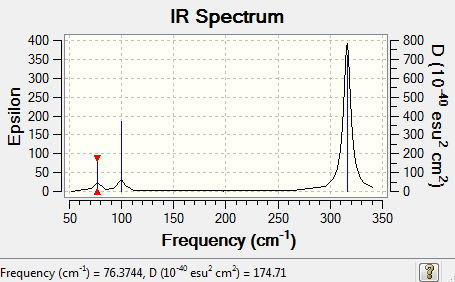
GaBr<sub>3</sub> Frequency Low frequencies --- -0.5252 -0.5247 -0.0024 -0.0010 0.0235 1.2010 Low frequencies --- 76.3744 76.3753 99.6982
GaBr3 Vibration
GaBr3_IRspectrum
BBr3
BBr3 optimisation
| File Name | SW_BBr3_OPT_GEN | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | Gen | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -64.43644666 | a.u. |
| RMS Gradient Norm | 0.00001415 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0003 | Debye |
| Point Group | CS | |
| Job cpu time: 0 days 0 hours 0 minutes 15.0 seconds. | ||
| Optimised Bond Length | 1.93, 1.93, 1.93 | Å |
| Literature Bond Lengthlit 2 | 1.893 | Å |
| Optimised Bond Angle | 120.0, 120.0, 120.0 |
BBr3 optimisation
Item Value Threshold Converged?
Maximum Force 0.000026 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000131 0.001800 YES
RMS Displacement 0.000065 0.001200 YES
Predicted change in Energy=-3.258105D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.9339 -DE/DX = 0.0 !
! A1 A(2,1,3) 119.9975 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0005 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.002 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Comparisons
| Compounds | Optimised Bond Length | Mean Bond Length | |
|---|---|---|---|
| BH3 | 1.19, 1.19, 1.19 | 1.19 | Å |
| GaBr3 | 2.35, 2.35, 2.35 | 2.35 | Å |
| BBr3 | 1.93, 1.93, 1.93 | 1.93 | Å |
Comments
What difference does changing the ligand have?
How are H and Br similar, how are they different?
What difference does changing the central element make?
How are B and Ga similar, how are they different?''
There are mainly four aspects: Change of overlap, change of atom size, change of polarity, change of the electronegativity.
Comparing the bond length of BH3 and BBr3, BBr bond is 0.74 Å longer than BH bond. The replacement of the H to Br group leads a poorer overlap between the ligand and boron, because the H has the s orbital overlap while the Br has the p orbital overlap. More electron density in Br increases the size of the atom due to stronger electrostatic repulsion as well as its electronegativity. The electronegativity of H, B and Br are 2.300, 2.051 and 2.685 lit 1 respectively. Therefore the polarity of the molecules is bigger in BBr3. Overall it gives a better interaction in BH3 than BBr3, hence a shorter bond appears in BH3.
The bond length of GaBr3 is 0.42 longer than BBr3. Both of Ga and B are in group 13, so their frontier orbitals are both p orbital. However Ga is a transition metal and B is electron deficient. So the 4p orbital in Ga is more deffused. As the group is going down, the size of the central element goes up as a consequence of the poorer overlap between the central atom and ligands. Ga is also more electropositive (1.756 lit 1) than B. So the bigger polarity leads to a weaker overlap and gives a longer bond.
A chemical bond is the electrostatic interaction between two opposite charged atoms.
Some strong interactions (common bonds) are identified as the covalent bonds (the pair of electrons are shared between two atoms), dative covalent bond (the electron pair is donated from one atom), the ionic bond (the Coulomb Force between a positive charged atom and a negative charged atom and the metallic bond (the delocalised electrons in the metal lattices).
Some weak interactions also can be found between molecules, such as London dispersion force, dipole-dipole interaction and hydrogen bond.
The bonds can be presented by the frontier orbital electrons. It valence bond theory and the molecular bond theory. So there are also single bond, double bond, triple bond, aromatic bond, etc. lit 3
In some structures, it doesn't mean there is no bond although Gaussview doesn't draw it out, although there is no bond drawn in the first few structures. It is because the bonds are presented based on a distance criteria in Gaussview. So no bond shown doesn't mean no bonds, but the bonds exceed the pre-defined distance value. There interaction is still there.
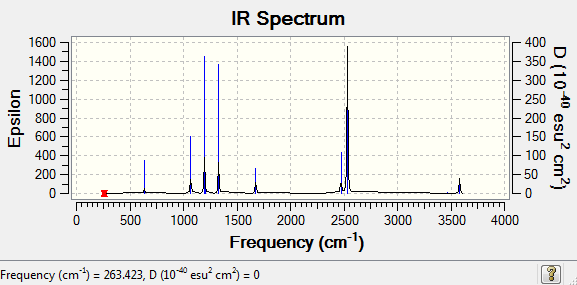
Vibrational frequencies of BH3 and GaBr3
| No. | Frequency of BH3 | Frequency of GaBr3 |
|---|---|---|
| 1 | 1163 | 76 |
| 2 | 1213 | 76 |
| 3 | 1213 | 100 |
| 4 | 2583 | 197 |
| 5 | 2716 | 316 |
| 6 | 2716 | 316 |
The higher frequency means the higher in energy. Therefore, it implies that the BH3 vibration is much higher in energy than GaBr3. It is compliance with the bonds lengths as a shorter bond distance is observed in B-H than in Ga-Br. It shows a higher interaction between B-H than Ga-Br. Ga-Br is weakly bonded.
There are six vibration modes in both of the molecules, because they have got the same symmetry and the same number of atoms in the molecule. However in BH3, it is mode 2&3 and 5&6 (degenerate) overlapping (single peak) to each other and mode 4 is zero in intensity. But in GaBr3, it is mode 1&2 and 5&6 (degenerate) overlapping (single peak) and mode 4 is zero in intensity.
The consequence of the observation of A2&E' modes/ A1'&E'modes lying close to each other but the latter is higher in energy is due to an orbital mixing occurs in GaBr3 but not in BH3.
Method and basis set have to be strickly the same to allow the comparison (even the keyword has to be the same). There is no worth comparing when the basic setting is different.
The purpose of carrying out a frequency calculation is to compare it with the real value obtained from lab to see the difference and the compliance. If two values match to each other, that's great. Otherwise it may either be something wrong with the experiment or something interesting on going.
Low frequency represents the translational an rotational degrees of freedom.
NH3
NH3 optimisation
| File Name | SW_BBr3_OPT_GEN | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776863 | a.u. |
| RMS Gradient Norm | 0.00000289 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.8464 | Debye |
| Point Group | C3V | |
| Job cpu time: 0 days 0 hours 0 minutes 10.0 seconds. | ||
| Optimised Bond Length | 1.02, 1.02, 1.02 | Å |
| Literature Bond Length*** | 2.35018 | Å |
| Optimised Bond Angle | 106, 106, 106 | |
| Literature Bond Angle*** | 2.35018 | |
| Reference*** | 2.35018 |
NH3 optimisation
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000010 0.001800 YES
RMS Displacement 0.000007 0.001200 YES
Predicted change in Energy=-7.830455D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7463 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7463 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7463 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.867 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3 Frequency
| File Name | SW_NH3_freq2 | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776873 | a.u. |
| RMS Gradient Norm | 0.00000028 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 1.8465 | Debye |
| Point Group | C3V | |
| Job cpu time: 0 days 0 hours 0 minutes 4.0 seconds. |
NH3 Frequency Low frequencies --- -6.1415 -4.4910 -4.4904 0.0014 0.0035 0.0150 Low frequencies --- 1089.3562 1693.9270 1693.9271
NH3 MO
| File Name | SW_NH3_MO | |
|---|---|---|
| File Type | .chk | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776863 | a.u. |
| RMS Gradient Norm | 0.00000000 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 1.8464 | Debye |
| Point Group | C3V | |
| Job cpu time: 0 days 0 hours 0 minutes 6.0 seconds. |
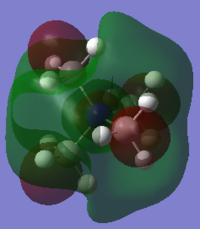
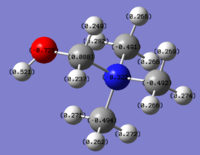
NH3 NBO Analysis
| NH3 NBO Analysis | Comment | |
|---|---|---|
 |
charge distributionColour range -1.000 to 1.000 | |
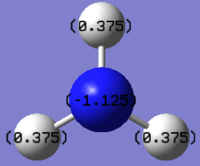 |
NBO charges for the nitrogen: -1.125 and hydrogen atom: 0.375 |
NH3BH3
NH3BH3 Optimisation
| File Name | SW_NH3BH3_OPT | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22468910 | a.u. |
| RMS Gradient Norm | 0.00000031 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 5.5646 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 0 minutes 25.0 seconds. |
NH<sub>3</sub>BH<sub>3</sub> Optimisation
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000005 0.000006 YES
RMS Displacement 0.000002 0.000004 YES
Predicted change in Energy=-3.646599D-12
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0185 -DE/DX = 0.0 !
! R2 R(2,7) 1.0185 -DE/DX = 0.0 !
! R3 R(3,7) 1.0185 -DE/DX = 0.0 !
! R4 R(4,8) 1.2098 -DE/DX = 0.0 !
! R5 R(5,8) 1.2098 -DE/DX = 0.0 !
! R6 R(6,8) 1.2098 -DE/DX = 0.0 !
! R7 R(7,8) 1.6677 -DE/DX = 0.0 !
! A1 A(1,7,2) 107.876 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.876 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.023 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.876 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0227 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0228 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.8738 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8739 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.5973 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.8739 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.5975 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.5974 -DE/DX = 0.0 !
! D1 D(1,7,8,4) -179.9991 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9992 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0009 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9991 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0008 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -179.9991 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.0008 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9992 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.9991 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3BH3 Frequency
| File Name | SW_NH3BH3_freq | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Change | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22468906 | a.u. |
| RMS Gradient Norm | 0.00000024 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 5.5646 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 0 minutes 28.0 seconds. |
NH3BH3 Frequency Low frequencies --- -3.1658 -2.5379 0.0014 0.0015 0.0015 1.8917 Low frequencies --- 263.4226 632.9765 638.4394
NH3BH3
Association Energy
Association Energy of NH3+BH3->NH3BH3
Basis Set: 6-31G(d,p)
E(NH3)= -56.55776863 a.u.
E(BH3)= -26.61532364 a.u.
E(NH3BH3)= -83.22468906 a.u.
then still in AU compute the energy difference and report this on your wiki ΔE=E(NH3BH3)-[E(NH3)+E(BH3)] = -83.22468906-[(-26.61532364)+(-56.55776863)]= -0.05159679 a.u.
the final step is to convert the energy difference from AU to kJ/mol
1 a.u. = 1 hartree = 2625.50 KJ/mol
ΔE=-0.0516 a.u. *2625.50 kJ/mol = -135.47 = KJ/mol
So the dissociation energy is +135.47 KJ/mol, endothermic dissociation. this number tells us the association energy for combinging a molecule of NH3 with one of BH3, conversely it is also the dissociation energy!
Week 2: Project: Ionic Liquids: Designer Solvents
Part 1: Comparison of selected 'onium' cations'
Optimisation and frequency of [N(CH3)4]+, [P(CH3)4]+ and [S(CH3)4]+
| onium cations | [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)4]+ |
|---|---|---|---|
| File Name | SW_[N(CH3)4]+_OPT_freq | SW_[P(CH3)4]+_OPT_freq3 | SW_[S(CH3)3]+_OPT_freq |
| File Type | .log | .log | .log |
| Calculation Type | FREQ | FREQ | FREQ |
| Calculation Method | RB3LYP | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) |
| Change | 1 | 1 | 1 |
| Spin | Singlet | Singlet | Singlet |
| E(RB3LYP) (a.u.) | -214.18127521 | -500.82700333 | -517.68325777 |
| RMS Gradient Norm (a.u.) | 0.00004006 | 0.00001637 | 0.00004777 |
| Imaginary Freq | 0 | 0 | 0 |
| Dipole Moment (Debye) | 0.0004 | 2.8852 | 0.9653 |
| Point Group | C1 | C1 | C1 |
| Job cpu time | 7 minutes 31.2 seconds. | 7 minutes 12.1 seconds. | 3 minutes 39.2 seconds. |
| IR Spectrum | 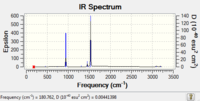 |
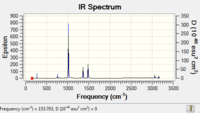 |
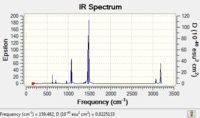
|
| Log file | [[N(CH3)4+ ]] | [[P(CH3)4+ ]] | [[S(CH3)3+ ]] |
| D Space file | [N(CH3)4+]] | [[P(CH3)4+]] | [[S(CH3)3+]] |
Optimisation of [N(CH3)4]+
Item Value Threshold Converged?
Maximum Force 0.000111 0.000450 YES
RMS Force 0.000028 0.000300 YES
Maximum Displacement 0.001413 0.001800 YES
RMS Displacement 0.000314 0.001200 YES
Predicted change in Energy=-1.051115D-07
Optimization completed.
-- Stationary point found.
Frequency of [N(CH3)4]+
Low frequencies --- -19.1188 -9.8202 -0.0009 -0.0006 -0.0001 21.7729 Low frequencies --- 180.8192 277.3212 288.7343
Optimisation of [P(CH3)4]+
Item Value Threshold Converged?
Maximum Force 0.000031 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000856 0.001800 YES
RMS Displacement 0.000253 0.001200 YES
Predicted change in Energy=-3.411912D-08
Optimization completed.
-- Stationary point found.
Frequency of [P(CH3)4]+
Low frequencies --- -18.0294 -4.5198 -0.0036 -0.0032 0.0009 14.5481 Low frequencies --- 153.7212 183.3695 191.3300
Optimisation of [S(CH3)3]+
Item Value Threshold Converged?
Maximum Force 0.000158 0.000450 YES
RMS Force 0.000050 0.000300 YES
Maximum Displacement 0.000943 0.001800 YES
RMS Displacement 0.000280 0.001200 YES
Predicted change in Energy=-2.620616D-07
Optimization completed.
-- Stationary point found.
Frequency of [S(CH3)3]+
Low frequencies --- -29.1030 -19.8918 -0.0034 -0.0031 -0.0024 11.1429 Low frequencies --- 159.6295 192.4985 205.8541
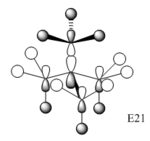
Molecular Orbital of [N(CH3)4]+, [P(CH3)4]+ and [S(CH3)4]+
| onium cations | [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)4]+ |
|---|---|---|---|
| File Name | SW_[N(CH3)4]+_MO | SW_[P(CH3)4]+_MO | SW_[S(CH3)3]+_MO |
| File Type | .log | .log | .log |
| Calculation Type | SP | SP | SP |
| Calculation Method | RB3LYP | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d,p) | 6-31G(d,p) | 6-31G(d,p) |
| Change | 1 | 1 | 1 |
| Spin | Singlet | Singlet | Singlet |
| E(RB3LYP) (a.u.) | -214.18127521 | -500.82698242 | -517.68325777 |
| RMS Gradient Norm (a.u.) | n/a | n/a | n/a |
| Imaginary Freq | n/a | n/a | n/a |
| Dipole Moment (Debye) | 0.0004 | 0.0021 | 0.9653 |
| Point Group | C1 | C1 | C1 |
| Job cpu time | 0 minutes 52.5 seconds | 1 minutes 15.4 seconds | 0 minutes 36.5 seconds |
| Image |  |
 |
|
| General Structure | tetrahedral | tetrahedral | pyramidal |
| Bond length (C-X) Å | 1.51 | 1.82 | 1.82 |
| Litt Bond length (C-X) | for N(CH3)4+N3-:1.47 to 1.53 | ||
| Bond angle (C-X-C) | 109.5 | 109.5 | 102.8 |
| Litt Bond angle | |||
| D Space file | [N(CH3)4+]] | [[P(CH3)4+]] | [[S(CH3)3+]] |
Comment on geometries
The bond length increases as the central nitrogen atom is replaced by phosphorus and sulfur. It indicates less bonding interaction and more diffused orbitals. In tetramethylammonium the bond length is within the range of the literature value. The literature value is influence in the presence of N3- group. lit 4
The bond angles remain the same at 109.5 degrees for N and P atoms. But it decreases to 102.8 degrees due to the lone pair repulsion on the sulfur atom.
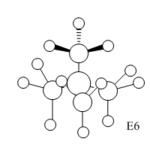
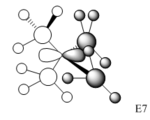
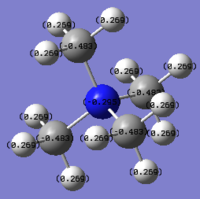
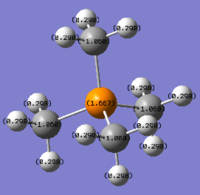
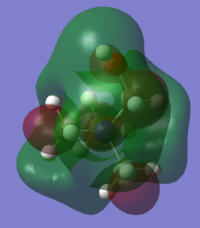
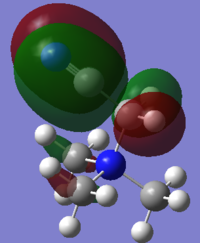
Charge Distribution Analysis
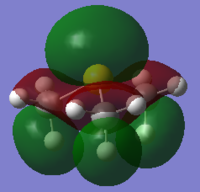
In sulfur cation, there is a slight difference in charge distribution between axial and equatorial hydrogen atoms. It is due to the H atoms are in the difference phase at its HOMO orbital.
| Bond C-X | C-N | C-P | C-S |
|---|---|---|---|
| contribution of X | 66.4%, s(25.0%), p(75.0%) | 40.4%, s(25.0%), p(74.1%) | 51.3%, s(17.0%), p(83.4%) |
| Contribution of C | 33.7%, s(20.8%), p(79.1%) | 59.6%, s(25.2%), p(74.7%) | 48.7%, s(19.7%), p(80.1%) |
Comment
Nitrogen contributes the most in comparison to the other two heteratoms in their C-X bonds with 66.4% and the phosphorus contributes the least. It matches the charge distribution of where nitrogen is the most electronegative and phosphorus is the most electropositive. In C-N, the nitrogen is sp3 hybridised, but there is more interaction with carbon p orbital than its s orbital. In C-P, both are sp3 hybrids bonded to each other as it is 25% contribution from s orbital and 75% contribution from p orbital. In C-S, approximately same contribution from two atoms, the about 20% contribution from s orbital and 80% from p orbital.
[NR4]+ According to the charge distribution just analysed, the nitrogen is actually negatively charged, although it is less negative than carbon atom. The positive charge is actually located on its hydrogen atom.
Part 2: Influence of functional groups
Optimisation and frequency of [N(CH3)3(CH2OH)]+ and [N(CH3)3(CH2CN)]+
| cations | [N(CH3)3(CH2OH)]+ | [N(CH3)3(CH2CN)]+ |
|---|---|---|
| File Name | SW_[N(CH3)3(CH2OH)]+_OPT_freq_vtight | SW_[N(CH3)3(CH2CN)]+_OPT_freq_vtight |
| File Type | .log | .log |
| Calculation Type | FREQ | FREQ |
| Calculation Method | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d,p) | 6-31G(d,p) |
| Change | 1 | 1 |
| Spin | Singlet | Singlet |
| E(RB3LYP) (a.u.) | -289.39470636 | -306.39376141 |
| RMS Gradient Norm (a.u.) | 0.00000035 | 0.00000041 |
| Imaginary Freq | 0 | 0 |
| Dipole Moment (Debye) | 8.2355 | 8.6721 |
| Point Group | C1 | C1 |
| Job cpu time | 21 minutes 54.9 seconds. | 24 minutes 50.5 seconds. |
| IR Spectrum | 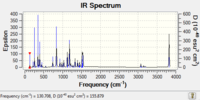 |
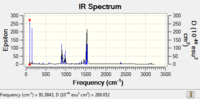
|
| Log file | [[N(CH3)3(CH2OH)+ ]] | [[N(CH3)3(CH2CN)+ ]] |
| D Space file | [[N(CH3)3(CH2OH)+]] | [[N(CH3)3(CH2CN)+]] |
Optimisation of [N(CH3)3(CH2OH)]+
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000005 0.000006 YES
RMS Displacement 0.000001 0.000004 YES
Predicted change in Energy=-2.728296D-13
Optimization completed.
-- Stationary point found.
Frequency of [N(CH3)3(CH2OH)]+
Low frequencies --- -9.6930 -7.6045 -4.7654 -0.0004 -0.0001 0.0005 Low frequencies --- 130.7079 213.6972 255.6339
Optimisation of [N(CH3)3(CH2CN)]+
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000003 0.000006 YES
RMS Displacement 0.000001 0.000004 YES
Predicted change in Energy=-3.560461D-13
Optimization completed.
-- Stationary point found.
Frequency of [N(CH3)3(CH2CN)]+
Low frequencies --- -4.7020 -4.2767 -0.0012 -0.0009 -0.0003 4.5253 Low frequencies --- 91.5853 153.8209 211.2666
Molecular Orbital of [N(CH3)3(CH2OH)]+ and [N(CH3)3(CH2CN)]+
| cations | [N(CH3)3(CH2OH)]+ | [N(CH3)3(CH2CN)]+ |
|---|---|---|
| File Name | SW_[N(CH3)3(CH2OH)]+MO | SW_[N(CH3)3(CH2CN)]+MO_vtight |
| File Type | .log | .log |
| Calculation Type | SP | SP |
| Calculation Method | RB3LYP | RB3LYP |
| Basis Set | 6-31G(d,p) | 6-31G(d,p) |
| Change | 1 | 1 |
| Spin | Singlet | Singlet |
| E(RB3LYP) (a.u.) | -289.39471123 | -306.39377029 |
| RMS Gradient Norm (a.u.) | n/a | n/a |
| Imaginary Freq | n/a | n/a |
| Dipole Moment (Debye) | 2.1359 | 5.7642 |
| Point Group | C1 | C1 |
| Job cpu time | 1 minutes 25.5 seconds. | 1 minutes 38.8 seconds. |
| D Space file | [[N(CH3)3(CH2OH)+]] | [[N(CH3)3(CH2CN)+]] |
Comment:
how has the shape of the orbitals changed?
LUMO: In tetramethylammonium LUMO orbital is central symmetrical with one positive phase on the central N atom and other four positive phases on methyl groups. In the cation with OH group, there is an additional positive phase on oxygen. So there is an overlap between the oxygen, the Carbon of CH2 and the central nitrogen. It stabilises the LUMO energy level. In the cation with CN group, the positive phase density is getting bigger. The negative phase density goes down so the repulsion between two phases gets weaker. Thus the LUMO energy goes down as well.
HOMO: There are less bonding interactions between the phases from left to right as the lobe on the atoms are smaller. Therefore the HOMO energy goes up.
has the energy of these orbitals moved?
The HOMO energy level rises up and LUMO energy level goes down when the functional groups are introduced. The effect is more obvious on CN group cation than OH group cation.
has the HOMO-LUMO gap changed in size?
The HOMO-LUMO gap decreases from left to right (no functional group to electron donating group to electron withdrawing group).
what chemical impact could these changes have?
The chemical impact of these is that it is more reactive with functional groups on the onium cation. And electron withdrawing group has a bigger influence than the electron donating group.
Charge Distribution Analysis
Bond length:
in alcohol: CH2-OH 1.37, CH2-N 1.55 CH3-N 1.50
in cyanide: CH2-CN 1.46, CH2-N 1.53 CH3-N 1.51
Reference
1. L. C. Allen, J. Am. Chem. Soc., 1989, 111:9003
2. Kagaku Benran, CRC Handbook of Chemistry and Physics, 87th Edition, 1984, 2, 649–661
3. G. N. Lewis, J. Am. Chem. Soc., 1916, 38(4), 762-785 [1]
4. K. O. Christe, W. W. Wilson, R. Bau and S. W. Bunte, J. Am. Chem. Soc., 1992, 114, 3411-3414 [2]