Rep:Mod:vino comp
NH3 Molecule
Optimisation
Using GaussView, the molecule NH3 was optimised, the results of this are shown in the table below:
| Molecule | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -56.44397188 a.u. |
| RMS Gradient | 0.00032440 a.u. |
| Point Group | C3V |
The optimised H-N-H bond angle was found to be 105.741°
| Item | Value | Threshold | Converged? |
| Maximum Force | 0.000004 | 0.000450 | YES |
| RMS Force | 0.000004 | 0.000300 | YES |
| Maximum Displacement | 0.000072 | 0.001800 | YES |
| RMS Displacement | 0.000035 | 0.001200 | YES |
| Predicted change in Energy | -5.986283D-10 |
The LOG file from GaussView: File:Vino NH3 opft pop log.LOG
NH Molecule |
Frequency Analysis
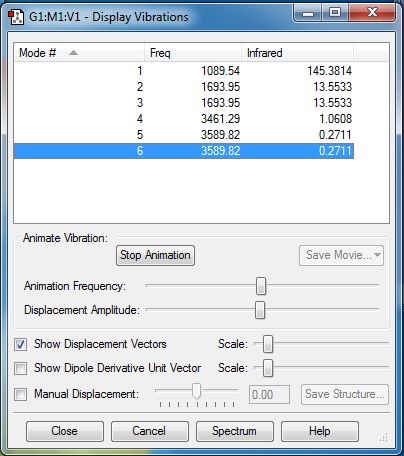
Display Vibrations Screenshot is shown below.
The 3N-6 rule would predict 6 modes of vibration. Modes 2 and 3, and modes 5 and 6 are degenerate as they have the same frequencies associated with them. Types of Vibrations Bending - Modes 1, 2 and 3 Bond Strecth - Modes 4, 5 and 6
Mode 4 is highly symmetric. Mode 1 is known as the umbrella mode. An experimental spectrum of gaseous ammonia would produce 3 bands.
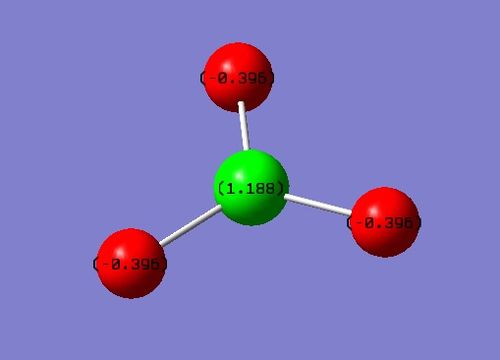
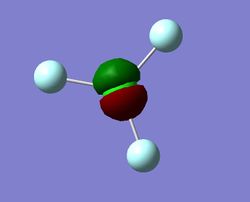
Charge Distribution
The above image shows the charges that would be predicted. The Nitrogen atom would have a negative charge as it is more electronegative, thus pulling the bonding electrons closer to itself; it gives it a negative charge. Conversely, the Hydrogen atom would have a positive charge.
N2 Molecule
Optimisation
| Molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient | 0.00000060 a.u. |
| Point Group | D∞h |
| Item | Value | Threshold | Converged? |
| Maximum Force | 0.000001 | 0.000450 | YES |
| RMS Force | 0.000001 | 0.000300 | YES |
| Maximum Displacement | 0.000000 | 0.001800 | YES |
| RMS Displacement | 0.000000 | 0.001200 | YES |
| Predicted change in Energy | -3.400985D-13 |
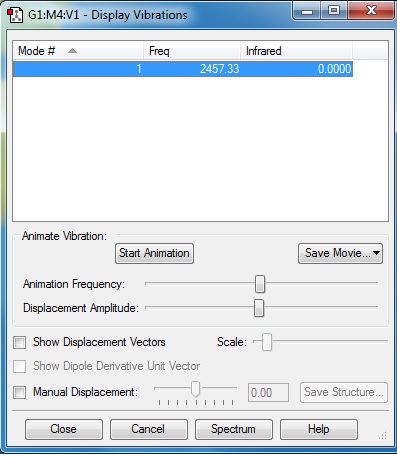
=Frequency Analysis
H2 Molecule
Optimisation
| Molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -1.15928020 a.u. |
| RMS Gradient | 0.09719500 a.u. |
| Point Group | D∞h |
| Item | Value | Threshold | Converged? |
| Maximum Force | 0.000000 | 0.000450 | YES |
| RMS Force | 0.000000 | 0.000300 | YES |
| Maximum Displacement | 0.000000 | 0.001800 | YES |
| RMS Displacement | 0.000001 | 0.001200 | YES |
| Predicted change in Energy | -1.164080D-13 |
Frequency Analysis
Harber-Bosch Process
| Energy / a.u. | Energy / kJ mol-1 | |
| E(NH3) | -56.44397188 a.u | -148,536.77 kJ mol-1 |
| 2*E(NH3) | -112.88794376 a.u. | -297,073.54 kJ mol-1 |
| E(N2) | -109.52412868 a.u. | -288,221.39 kJ mol-1 |
| E(H2) | -1.15928020 a.u. | -3,050.74 kJ mol-1 |
| 3*E(H2) | -3.4778406 a.u. | -9152.22 kJ mol-1 |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | 300.07 kJ/mol |
ClF3 Molecule
Optimisation
Using GaussView, the molecule ClF3 was optimised, the results of this are shown in the table below:
| Molecule | ClF3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d.p) |
| E(RB3LYP) | -759.44149573 a.u. |
| RMS Gradient | 0.00000553 a.u. |
| Point Group | D3H |
The optimised F-Cl-F bond angle is 120°
| Item | Value | Threshold | Converged? |
| Maximum Force | 0.000011 | 0.000450 | YES |
| RMS Force | 0.000007 | 0.000300 | YES |
| Maximum Displacement | 0.000049 | 0.001800 | YES |
| RMS Displacement | 0.000032 | 0.001200 | YES |
| Predicted change in Energy | -8.166595D-10 |
The LOG file from GaussView: File:VINO CLF3 OPTF POP.LOG
ClF Molecule |
Display Vibrations
Display Vibrations Screenshot is shown below.

Modes 1 and 2, and modes 5 and 6 are degenerate as they have the same frequencies. Mode 3 is th umbrella mode.
Charge Distribution
The above image shows the charges that would be predicted. The Chlorine atom w has a positive charge and the Fluorine atoms have a negative charge. Fluorine is highly electronegative, thus it attracts the bonding electrons closer to itself; hence, the negative charge. Conversely, the Chlorine atom has a positive charge.
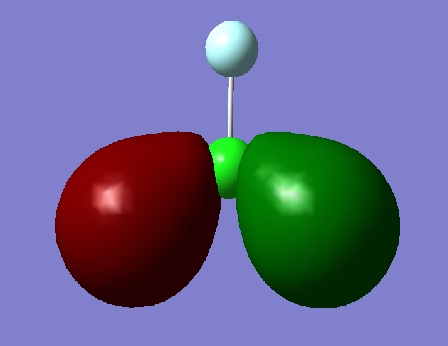
Oribtals
The image above shows the 5th Molecular Orbital. Of all the MO's shown here, this is the deepest in energy. The image shows 1s orbital in the Chlorine atom.
The image above shows the 7th Molecular Orbital. It is one of 3 degenerate MO's with an energy of -7.45 a.u. Again,this MO, a mixture of bonding and anti-bonding orbitals, is relatively deep in energy. This shows a sigma bond.
The image above shows the 10th Molecular Orbital.
The image above shows the 11th Molecular Orbital. Both the images above show MO's which are degenerate of one another. They both gave the same energies of -1.18 a.u. The 10th MO shows the interaction between 2s and 2s in Fluorine and Chlorine respectively as well as the interactions between 2s orbitals in two neighboring Florine atoms. The 11th MO shows the interaction between the orbitals in Chlorine and Fluorine. Again, this interaction produces bonding and anti-bonding molecular orbitals. The 2s orbital of 2 Florine atoms interact with the 2s orbital in the Chlorine atom.
The image above shows the 15th Molecular Orbital. This has an energy of -0.516 a.u. so it is still relatively deep in energy. It shows the interactions between the two 2p in each of the atoms. The bonding molecular orbital is filled, so the bonding is strong.