Rep:Mod:tryout
NH3 molecule
The molecule in question is ammonia
The calculation method is RB3LYP
The basis set is 6-31G(d.p.)
The final energy (E(RB3LYP)) is -56.55776873
The RMS gradient is 0.00000485
The point group of ammonia is C3V
The Bond distance N-H is 1.01798
The Bong angle is 105.741
Item box
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986279D-10
Optimization completed.
-- Stationary point found.
The optimisation file is here
gif of molecule
NH3 molecule |
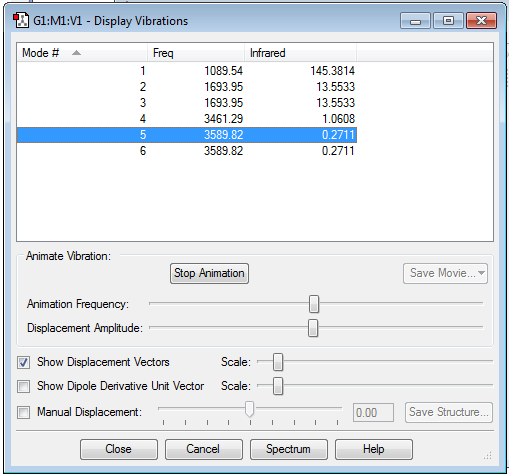
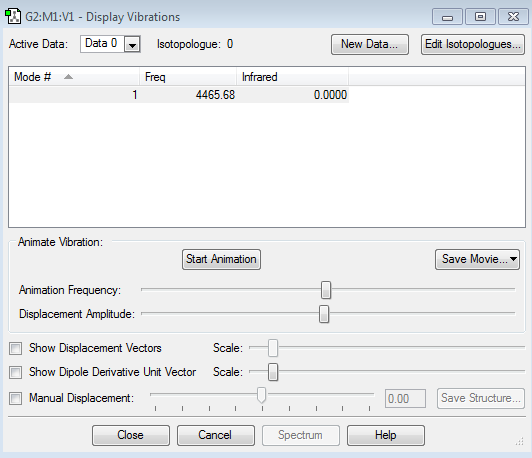
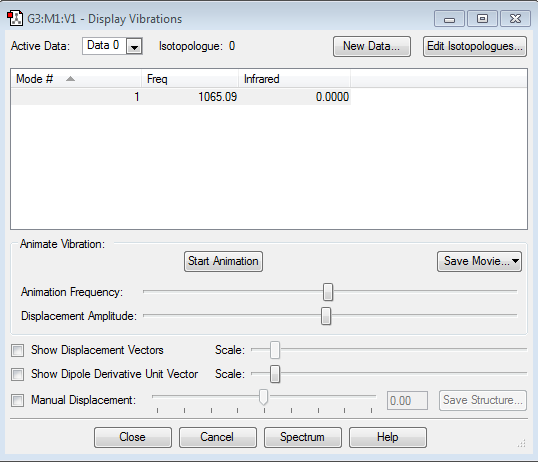
The mode we'd expect from the 3N-6 rule is 6, which is what the display vibrations shows us.
The 2nd and 3rd are degenerate as well as the 5th and 6th.
1st,2nd and 3rd vibrations are bending vibrations
4th, 5th, and 6th vibrations are stretching vibrations
The 4th mode is highly symmetric
The 1st mode is the umbrella mode
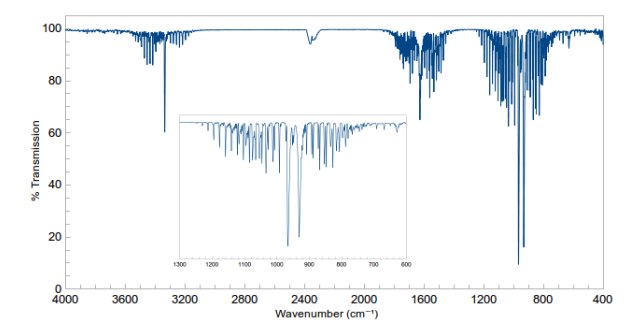
One would expect there to be 3 bands in the spectrum of gaseous ammonia. This is due to the 4th mode being symmetrical and so unchaging in polarity, and the other three modes changing in polarity. This is infact backed up by literature data. The graph shows three bans within this graph, as was predicted by the vibration values calculated in GuassView.
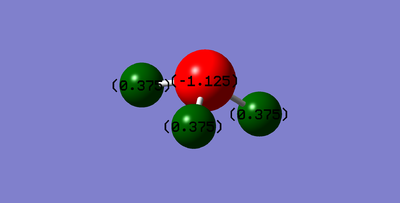
One would expect the Nitrogen to have a negative charge in comparison to the hydrogens. This is because it is highly electronegative (3.04), and so it has a tendency to pull electron density towards itself, and so create a negatively charged cloud around it. N has a charge of -1.125 and H has a charge of 0.375
N2 molecule
The molecule in question is nitrogen
The calculation method is RB3LYP
The basis set is 6-31G(D,P)
The Total energy is -109.52412868
The RMS gradient is 0.00000060
The point group of nitrogen is Dinfinityh
The Bond distance N-H is 1.10550
The Bong angle is 180
Item box
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401029D-13
Optimization completed.
-- Stationary point found.
The optimisation file is here
gif of molecule
N2 molecule |
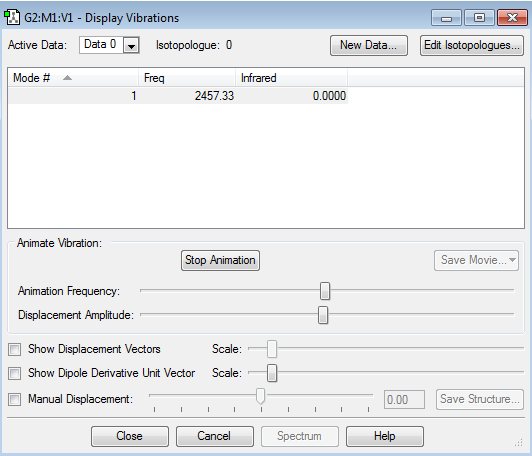
As can be seen in the table, there are no negative frequencies.
H2 molecule
The molecule in question is hydrogen
The calculation method is RB3LYP
The basis set is 6-31G(D,P)
The Total energy is -1.17853936
The RMS gradient is 0.00000017
The point group of hydrogen is Dinfinityh
The Bond distance H-H is 0.74279
The Bong angle is 180
Item box
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
The optimisation file is here
gif of molecule
H2 molecule |
As can be seen in the table, there are no negative frequencies.
reaction energy
We can determine the energy for the reaction of N2 + 3H2 -> 2NH3. In your wiki complete the items below for energies in atomic units:
E(NH3)= -56.55776873 = -148492.4218
2xE(NH3)= -113.1155375 = -296984.8436
E(N2)= -109.52412868 = -287555.5998
E(H2)= -1.17853936 = -3094.25509
3xE(H2)= -3.53561808 = -9282.765269
ΔE=2xE(NH3)-[E(N2)+3xE(H2)]= -0.05579074 = -146.478531
The literature value for the Haber Process is -92.4kJ/mol[2] which is a significantly different value. However, this is the value for when the temperature and pressure are optimised for the reaction - 400-450 degrees centigrade, 200 atm and an iron catalyst. The results calculated are for the molecules at standard conditions.
F2 molecule
The molecule in question is fluorine
The calculation method is RB3LYP
The basis set is 6-31G(D,P)
The Total energy is -199.49825218
The RMS gradient is 0.00007365
The point group of nitrogen is Dinfinityh
The Bond distance N-H is 1.40281
The Bong angle is 180
Item box
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995025D-08
Optimization completed.
-- Stationary point found.
The optimisation file is here
gif of molecule
F2 molecule |
As can be seen in the table, there are no negative frequencies. Furthermore, because F2 is a dihomonuclear molecule, it has no charge.
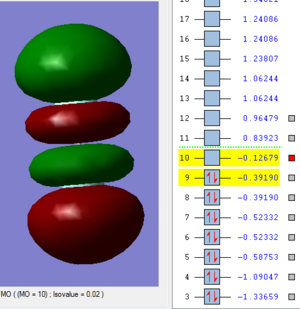
Molecular Orbitals
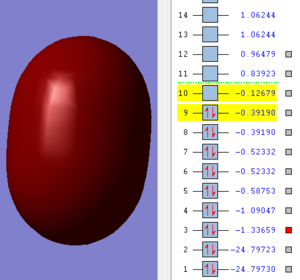
This image shows a sigma molecular orbital. It is made with the two 2s atomic orbitals of the fluorine atoms.
This image shows an antibonding molecular orbital. It is made with the two 2s atomic orbitals of the fluorine atoms.
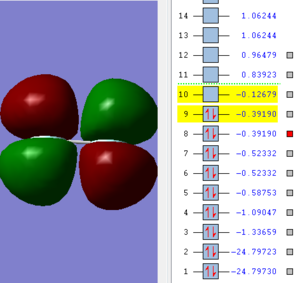
This image shows a pi molecular orbital. It is made with the two 2px atomic orbitals of the fluorine atoms.
This image shows an antibonding pi molecular orbital. It is made with the two 2px atomic orbitals of the fluorine atoms. This is the HOMO.
This image shows an antibonding pi molecular orbital. It is made with the two 2px atomic orbitals of the fluorine atoms. This is the LUMO.
Referencing
[1] Melville, J. Infrared Spectroscopy and Interferometry as Methods for Structural Determination of Ammonia. (1995).
[2] Clark, J. Case Study: The Haber Process. Chemistry LibreTexts (2017). Available at: https://chem.libretexts.org/Core/Physical_and_Theoretical_Chemistry/Equilibria/Case_Studies/Haber_Process. (Accessed: 3rd March 2017)