Rep:Mod:theKfiles
Inorganic week one
Days 1 & 2
Optimising BH3
BH3 was drawn in Gaussview then optimised using the B3LYP method and 3-21G basis set. The logfile can be found here: File:KWL BH3OPT.LOG and the convergence table and bond/angle values from it:
Item Value Threshold Converged?
Maximum Force 0.000220 0.000450 YES
RMS Force 0.000106 0.000300 YES
Maximum Displacement 0.000709 0.001800 YES
RMS Displacement 0.000447 0.001200 YES
Predicted change in Energy=-1.672423D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1947 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1948 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1944 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0157 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9983 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.986 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| File Name | KWL_BH3OPT |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 3-21G |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.46226429 au |
| RMS Gradient Norm | 0.00008851 au |
| Imaginary Freq | |
| Dipole Moment | 0.0003 Debye |
| Point Group | CS |
| Job cpu time | 33.0 seconds |
The optimised output molecule was then submitted for further optimisation using the more accurate 6-31G(d,p) basis set, with the results here: File:KWL BH3OPT6-31G.LOG
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000099 0.000300 YES
Maximum Displacement 0.000875 0.001800 YES
RMS Displacement 0.000418 0.001200 YES
Predicted change in Energy=-1.452073D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1926 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1928 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1924 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0146 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9988 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9866 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| File Name | KWL_BH3OPT6-31G |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61532358 au |
| RMS Gradient Norm | 0.00008206 au |
| Imaginary Freq | |
| Dipole Moment | 0.0003 Debye |
| Point Group | CS |
| Job cpu time | 38.0 seconds |
Optimising GaBr3
As GaBr3 has heavier atoms, pseudo potentials were used to optimise it. The LanL2LZ set was chosen, which uses the D95V basis set for first-row elements and Los Alamos ECP pseudo potentials for other elements. The molecule's symmetry was restricted to the D3h point group. This was submitted to the HPC service to give these results: DOI:10042/26066
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
Predicted change in Energy=-1.282693D-12
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.3502 -DE/DX = 0.0 !
! R2 R(1,3) 2.3502 -DE/DX = 0.0 !
! R3 R(1,4) 2.3502 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| File Name | KWL_GaBr3opt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -41.70082783 au |
| RMS Gradient Norm | 0.00000016 au |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | D3H |
| Job cpu time | 11.5 seconds |
Optimising BBr3
A mixture of methods was used for BBr3: 6-31G(d,p) for the boron atom and LanL2DZ for the bromine atoms. This was submitted to the HPC service to give these results: DOI:10042/26067
Item Value Threshold Converged?
Maximum Force 0.000020 0.000450 YES
RMS Force 0.000010 0.000300 YES
Maximum Displacement 0.000109 0.001800 YES
RMS Displacement 0.000059 0.001200 YES
Predicted change in Energy=-1.855277D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.9339 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0026 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0012 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9962 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| File Name | KWL_BBr3opt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | Gen |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -64.43645212 au |
| RMS Gradient Norm | 0.00000884 au |
| Imaginary Freq | |
| Dipole Moment | 0.0005 Debye |
| Point Group | CS |
| Job cpu time | 19.2 seconds |
Discussion
| Molecule | Bond length / Å |
|---|---|
| BH3 | 1.1926 |
| BBr3 | 1.9340 |
| GaBr3 | 2.3502 |
The bond length increases down this table due to the increase in atomic size: bromine's covalent radius 120±3 pm is far greater than hydrogen's of 35±5 pm. Similarly, gallium's of 122±3 pm is larger than boron at 84±3 pm. As B and Ga are in the same group, and as Br and H are both one electron from a full shell, the bonding is similar in all cases, thus atomic size is the largest contributor.
For some structures and early iterations of optimised molecules, GaussView does not show bonds. A bond is drawn only when two atoms are within a certain distance of each other as decided by GaussView; Gaussian itself has no concept of a bond, and considers only the position of the electrons. A bond is a human abstraction for when the majority of electron density between two atoms in a molecule is in a wavefunction that stabilises the energy compared to that of the free atoms, i.e. a bonding orbital.
Days 3 & 4
BH3 frequencies
The optimised BH3 molecule from above was submitted for frequency calculations in Gaussian; however, its highest low frequency was 43.7190cm-1, suggesting the molecule was not at a true minimum. The 6-31G(d,p) optimisation was repeated with additional keywords:
# opt=tight b3lyp/6-31g(d,p) geom=connectivity scf=conver=9 int=ultrafine
This gave these results: File:KWL BH3OPTTIGHT.LOG
| File Name | BH3OPTTIGHT |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61532364 au |
| RMS Gradient Norm | 0.00000211 au |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | D3H |
| Job cpu time | 5.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000004 0.000015 YES
RMS Force 0.000003 0.000010 YES
Maximum Displacement 0.000017 0.000060 YES
RMS Displacement 0.000011 0.000040 YES
Predicted change in Energy=-1.021890D-10
Optimization completed.
-- Stationary point found.
The frequency calculation was repeated. This gave the following results: File:KWL BH3FREQTIGHT.LOG
| File Name | BH3FREQTIGHT |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61532364 au |
| RMS Gradient Norm | 0.00000215 au |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | D3H |
| Job cpu time | 7.0 seconds |
Low frequencies --- -11.7222 -11.7144 -6.6058 0.0008 0.0278 0.4278 Low frequencies --- 1162.9743 1213.1388 1213.1390
And the following vibrations:
| No. | Form of vibration | Frequency / cm-1 | Intensity | Symmetry |
|---|---|---|---|---|
| 1 | All H atoms move 'up and down' in z-direction in concerted motion. The B atom moves in opposition along the z-axis to retain centre of mass. | 1163 | 93 | A2" |
| 2 | Two H atoms rock in plane with molecule in concerted motion; the third rocks across a far greater range along x-direction. The B atoms opposes the final H atom's motion along the x-axis. | 1213 | 14 | E' |
| 3 | Two H atoms move towards and away from each other in a scissoring motion. The B atom and third H atom move slightly along y-axis to oppose the scissoring. | 1213 | 14 | E' |
| 4 | All thee H atoms move radially in and out in concerted motion from the stationary B atom. | 2583 | 0 | A1' |
| 5 | Two H atoms move in and out radially in opposing motion. The B atom moves slightly along the x-axis | 2716 | 126 | E' |
| 6 | Two H atoms move in and out radially in concerted motion, the third moves a greater amount to oppose them radially along the y-axis. The B atom opposes this motion slightly along the y-axis. | 2716 | 126 | E' |
Although there are 6 vibrations, there are two degenerate pairs (2,3 and 5,6) and vibration 4 causes no change in dipole moment, so is not IR-active. Thus, only three peaks can be seen on the spectrum:
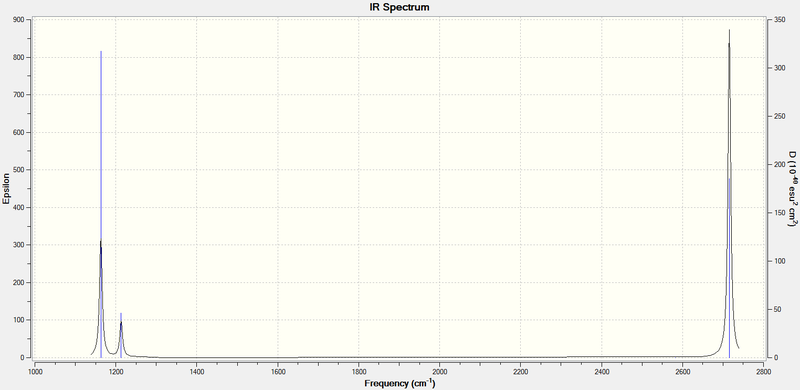
GaBr3 frequency analysis
The optimised GaBr3 molecule was submitted for frequency calculations, to give the following results: DOI:10042/26187
| File Name | GaBr3FREQ |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | LANL2DZ |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -41.70082783 au |
| RMS Gradient Norm | 0.00000011 au |
| Imaginary Freq | 0 |
| Dipole Moment | 0.0000 Debye |
| Point Group | D3H |
| Job cpu time | 8.2 seconds |
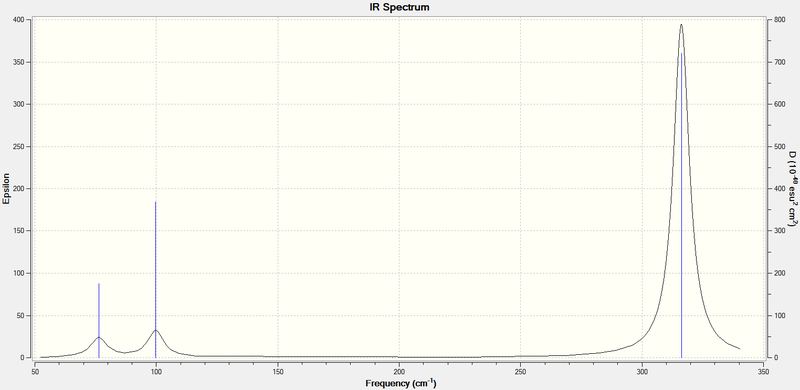
Low frequencies --- -0.5252 -0.5247 -0.0024 -0.0010 0.0235 1.2010 Low frequencies --- 76.3744 76.3753 99.6982
And this IR spectrum:
| Vibration | a2" | e' | e' | a1' | e' | e' |
|---|---|---|---|---|---|---|
| BH3 frequency | 1163 | 1213 | 1213 | 2583 | 2716 | 2716 |
| BH3 intensity | 93 | 14 | 14 | 0 | 126 | 126 |
| GaBr3 frequency | 100 | 76 | 76 | 197 | 316 | 316 |
| GaBr3 intensity | 9 | 3 | 3 | 0 | 57 | 57 |
The low frequencies, which represent the vibrations of the molecule's centre of mass, are very close to zero, which shows that the GaBr3 molecule is well-optimised to an energy minimum. The a2" vibration had been re-ordered to number 4; however, in the table above it is shown first, for easy comparison with the BH3 frequencies.
The higher atomic masses in GaBr3 are reflected in the much lower frequencies of vibration, as is to be expected from the equation for frequency of simple harmonic motion:
In GaBr3 the Ga atom moves more than the Br atoms; whereas in BH3 it is the H atoms which move the most.
The vibrations are split into three at low energies, which correspond to bends, and three at much higher energies which correspond to stretches; this is a common sight on IR spectra, where C-H and O-H stretches are usually the highest-energy vibrations visible.
BH3 molecular orbitals
The optimised BH3 molecule was submitted to Gaussian for molecular orbital calculations using the following keywords:
# b3lyp/6-31g(d,p) pop=(nbo,full) geom=connectivity int=ultrafine scf=conver=9
To give these results: File:KWL BH3POPULATION.LOG
| File Name | BH3population |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -26.61532364 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | D3H |
| Job cpu time | 2.0 seconds |
A comparison of the calculated MOs and theoretical ones is shown below:

The computed MOs are fit the shape of the theoretical ones, with two notable differences: all computed e' are asymmetrical, and the computed antibonding a1' orbital shape suggests that contribution from the boron atomic orbital is not larger than the contributions from the hydrogen atomic orbitals.
NH3 natural bond orbitals
A molecule of NH3 was optimised with the tight criteria as used for BH3 to give these results: File:KWL NH3OPTTIGHT.LOG
| File Name | NH3OPTTIGHT |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -56.55776873 au |
| RMS Gradient Norm | 0.00000323 au |
| Imaginary Freq | |
| Dipole Moment | 1.8465 Debye |
| Point Group | C3V |
| Job cpu time | 12.0 seconds |
Maximum Force 0.000006 0.000015 YES
RMS Force 0.000004 0.000010 YES
Maximum Displacement 0.000012 0.000060 YES
RMS Displacement 0.000008 0.000040 YES
Predicted change in Energy=-9.845951D-11
Optimization completed.
-- Stationary point found.
A frequency analysis was then carried out to check the optimisation: File:KWL NH3FREQTIGHT.LOG
| File Name | NH3FREQTIGHT |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -56.55776873 au |
| RMS Gradient Norm | 0.00000322 au |
| Imaginary Freq | |
| Dipole Moment | 1.8465 Debye |
| Point Group | C3 |
| Job cpu time | 12.0 seconds |
Low frequencies --- -0.0130 -0.0024 0.0010 7.0758 8.0923 8.0926 Low frequencies --- 1089.3840 1693.9368 1693.9368
Finally, a population analysis was run: File:KWL NH3POPULATION.LOG

| File Name | NH3POPULATION |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -56.55776873 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 1.8465 Debye |
| Point Group | C3V |
| Job cpu time | 3.0 seconds |
This gave the NBO charge-coloured image as seen on the right, with a colour range from -1.125 to 1.125. Absolute charges on the atoms were -1.125 on the N atom and +0.375 on each H atom.
Ammonia borane association energy
Though the absolute values of energy that Gaussian calculates are not relatable to any real thermodynamic property, energy differences between similar molecules, if the same method and basis set have been used, can be compared. A molecule of NH3BH3 was drawn in GaussView and optimised to give the following results:
| File Name | NH3OBH3OPT |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 0 |
| Spin | Singlet |
| E(RB3LYP) | -83.22468907 au |
| RMS Gradient Norm | 0.00000129 au |
| Imaginary Freq | |
| Dipole Moment | 5.5646 Debye |
| Point Group | C1 |
| Job cpu time | 56.0 seconds |
Item Value Threshold Converged?
Maximum Force 0.000002 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000032 0.000060 YES
RMS Displacement 0.000010 0.000040 YES
Predicted change in Energy=-1.066426D-10
Optimization completed.
-- Stationary point found.
This energy was compared with that of the tightly-optimised BH3 and NH3 molecules above to find the association energy by:
ΔE = E(NH3BH3) - (E(NH3) + E(BH3))
| E(NH3) | -56.55776873 au |
| E(BH3) | -26.61532360 au |
| E(NH3BH3) | -83.22468910 au |
| ΔE | -0.05159677 au |
The association energy given above in Hartrees is equivalent to -135.47 kJmol-1. This is a reasonable answer; for comparison, the N-N bond energy is about 170kJmol-1.
Ionic solvents mini-project
Introduction
Using the methods learnt above, three methyl 'onium' cations ([N(CH3)4]+ [P(CH3)4]+ and [S(CH3)3]+) were analysed and compared in terms of their structure and charge distribution. Several molecular orbitals of [N(CH3)4]+ were analysed in detail and the effect of adding a nitrile or alcohol functional group was investigated.
Method
The three cations [N(CH3)4]+ [P(CH3)4]+ and [S(CH3)3]+ were drawn in GaussView with broken symmetry, then submitted to Gaussian on the HPC cluster for optimisation using the RB3LYP method and 6-31G(d,p) basis set. Extra criteria for more-accurate optimisation were defined to ensure minimal low frequencies, thus the full keywords used were:
# opt=tight b3lyp/6-31g(d,p) geom=connectivity scf=conver=9 int=ultrafine
The optimised molecules were then submitted for frequency analysis and full NBO population analysis. The optimised [N(CH3)4]+ molecule was modified to make both [N(CH3)3(CH2OH)]+ and [N(CH3)3(CH2CN)]+, which were submitted for optimisation, frequency and population analysis as above.
Results
[N(CH3)4]+
Optimisation
| File Name | ammoniumopt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -214.18127320 au |
| RMS Gradient Norm | 0.00000464 au |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | C1 |
| Job cpu time | 49 min 7.4 sec |
Item Value Threshold Converged?
Maximum Force 0.000011 0.000015 YES
RMS Force 0.000003 0.000010 YES
Maximum Displacement 0.000060 0.000060 YES
RMS Displacement 0.000019 0.000040 YES
Predicted change in Energy=-2.726737D-10
Optimization completed.
-- Stationary point found.
Frequency analysis
| File Name | ammoniumfreq |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -214.18127270 au |
| RMS Gradient Norm | 0.00000431 au |
| Imaginary Freq | 0 |
| Dipole Moment | 0.0000 Debye |
| Point Group | C1 |
| Job cpu time | 19 min 5.1 sec |
Low frequencies --- -12.5751 -5.9636 -0.0008 -0.0003 0.0010 10.3939 Low frequencies --- 182.2506 287.9690 288.3252
Population analysis
{DOI|10042/26262}}
| File Name | ammoniumpop |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -214.18127270 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 0.0000 Debye |
| Point Group | C1 |
| Job cpu time | 2 min 2.1 sec |
[P(CH3)4]+
Optimisation
Item Value Threshold Converged?
Maximum Force 0.000015 0.000015 NO
RMS Force 0.000004 0.000010 YES
Maximum Displacement 0.005153 0.000060 NO
RMS Displacement 0.001275 0.000040 NO
Predicted change in Energy=-2.841724D-08
Optimization stopped.
-- Number of steps exceeded, NStep= 92
-- Flag reset to prevent archiving.
This partially-optimised molecule was re-submitted: DOI:10042/26223
| File Name | phosphoniumopt2 |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -500.82701141 au |
| RMS Gradient Norm | 0.00000494 au |
| Imaginary Freq | |
| Dipole Moment | 0.0002 Debye |
| Point Group | C1 |
| Job cpu time | 56 min 24.6 sec |
Item Value Threshold Converged?
Maximum Force 0.000013 0.000015 YES
RMS Force 0.000004 0.000010 YES
Maximum Displacement 0.000055 0.000060 YES
RMS Displacement 0.000017 0.000040 YES
Predicted change in Energy=-3.244399D-06
Optimization completed.
-- Stationary point found.
Frequency analysis
| File Name | phosphoniumfreq |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -500.82701139 au |
| RMS Gradient Norm | 0.00000507 au |
| Imaginary Freq | 0 |
| Dipole Moment | 0.0002 Debye |
| Point Group | C1 |
| Job cpu time | 17 min 25.6 sec |
Low frequencies --- -9.3532 0.0015 0.0020 0.0029 7.5058 8.8354 Low frequencies --- 155.9719 191.5135 191.9097
Population analysis
| File Name | phosphoniumpop |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -500.82701141 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 0.0002 Debye |
| Point Group | C1 |
| Job cpu time | 1 min 58.8 sec |
[S(CH3)3]+
Optimisation
| File Name | sulfoniumopt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -517.68327476 au |
| RMS Gradient Norm | 0.00000408 au |
| Imaginary Freq | |
| Dipole Moment | 0.9651 Debye |
| Point Group | C1 |
| Job cpu time | 36 min 51.2 sec |
Item Value Threshold Converged?
Maximum Force 0.000011 0.000015 YES
RMS Force 0.000003 0.000010 YES
Maximum Displacement 0.000017 0.000060 YES
RMS Displacement 0.000006 0.000040 YES
Predicted change in Energy=-1.166255D-06
Optimization completed.
-- Stationary point found.
Frequency analysis
| File Name | sulfoniumfreq |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -517.68327474 au |
| RMS Gradient Norm | 0.00000415 au |
| Imaginary Freq | 0 |
| Dipole Moment | 0.9651 Debye |
| Point Group | C1 |
| Job cpu time | 8 min 34.5 sec |
Low frequencies --- -8.3220 -3.1574 -0.0030 -0.0013 -0.0009 10.7753 Low frequencies --- 161.9116 199.3135 199.9844
Population analysis
| File Name | sulfoniumfreq |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -517.68327476 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 0.9651 Debye |
| Point Group | C1 |
| Job cpu time | 1 min 14.9 sec |
[N(CH3)3(CH2OH)]+
Optimisation
| File Name | CH2OHammoniumopt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -289.39470724 au |
| RMS Gradient Norm | 0.00000041 au |
| Imaginary Freq | |
| Dipole Moment | 2.1357 Debye |
| Point Group | C1 |
| Job cpu time | 2 hrs 1 min 28.5 sec |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000018 0.000060 YES
RMS Displacement 0.000005 0.000040 YES
Predicted change in Energy=-5.139318D-12
Optimization completed.
-- Stationary point found.
Frequency analysis
| File Name | CH2OHammoniumfreq |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -289.39470724 au |
| RMS Gradient Norm | 0.00000045 au |
| Imaginary Freq | 0 |
| Dipole Moment | 2.1357 Debye |
| Point Group | C1 |
| Job cpu time | 23 min 1.5 sec |
Low frequencies --- -8.4393 -5.0339 -1.1018 -0.0011 -0.0008 -0.0008 Low frequencies --- 131.1058 213.4629 255.7146
Population analysis
| File Name | CH2OHammoniumpop |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -289.39470724 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 2.1357 Debye |
| Point Group | C1 |
| Job cpu time | 2 min 22.8 sec |
[N(CH3)3(CH2CN)]+
Optimisation
| File Name | CH2CNammoniumopt |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -306.39376383 au |
| RMS Gradient Norm | 0.00000035 au |
| Imaginary Freq | |
| Dipole Moment | 5.7642 Debye |
| Point Group | C1 |
| Job cpu time | 24 min 50.1 sec |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000007 0.000060 YES
RMS Displacement 0.000001 0.000040 YES
Predicted change in Energy=-1.242860D-12
Optimization completed.
-- Stationary point found.
Frequency analysis
| File Name | CH2CNammoniumfreq |
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -306.39376383 au |
| RMS Gradient Norm | 0.00000037 au |
| Imaginary Freq | 0 |
| Dipole Moment | 5.7642 Debye |
| Point Group | C1 |
| Job cpu time | 25 min 52.3 sec |
Low frequencies --- -2.6294 -0.0013 -0.0011 -0.0007 7.1460 9.6655 Low frequencies --- 91.7741 154.0284 210.9285
Population analysis
| File Name | CH2OHammoniumpop |
| File Type | .log |
| Calculation Type | SP |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Charge | 1 |
| Spin | Singlet |
| E(RB3LYP) | -289.39470724 au |
| RMS Gradient Norm | |
| Imaginary Freq | |
| Dipole Moment | 2.1357 Debye |
| Point Group | C1 |
| Job cpu time | 2 min 22.8 sec |
Discussion
Geometries
| Molecule | CXC bond angle / ° | XCH bond angle / ° | HCH bond angle / ° | X-C bond lengths / Å | C-H bond lengths / Å |
|---|---|---|---|---|---|
| [N(CH3)4]+ | 109.5 | 108.9 | 110.0 | 1.51 | 1.09 |
| [P(CH3)4]+ | 109.5 | 109.9 | 109.0 | 1.82 | 1.09 |
| [S(CH3)3]+ | 102.7 | 107.3 & 110.6 | 109.4 & 111.1 | 1.82 | 1.09 |
The two tetramethyl cations have a tetrahedral arrangement as shown by the 109.5° central angles. Trimethylsulfonium has a trigonal pyramidal structure and a lower central bond angle of 102.7°. In VSEPR theory, this is rationalised by the existence of a lone pair in the fourth 'tetrahedral' position which is more repulsive than bonding pairs, thus the methyl groups are pushed closer together by this repulsion.
The X-C bond lengths reflect the covalent radii of the central atom: P (106pm) and S (102pm) are very similar, but both much larger than N (75pm). The C-H bond lengths in all the cations are unchanged from those in methane; however, the structure of the methyl groups is not perfectly tetrahedral. In [N(CH3)4]+ the hydrogen atoms are attracted slightly towards the N atom as shown by the XCH angle being less than the tetrahedral angle; in [P(CH3)4]+ they are pushed slightly away. This could be due to an attraction between the H atoms and the central atom, but which has a more distant minimum for phosphorous due to its larger, more-diffuse orbitals. In [S(CH3)3]+ not all hydrogen atoms are equivalent: on each methyl group, two H atoms point 'up' close to the central atom while the third one points 'down' in the opposite direction to the S lone pair. The differing angles for the two 'up' and the 'down' H atoms are shown respectively on the table above. The two 'up' H atoms are closer to each other than the tetrahedral angle, and are attracted closer to the central atom, whereas the 'down' hydrogen is further away. As shown below the H atoms have a slight positive charge, thus the 'up' atoms may be attracted to the S lone pair.
[N(CH3)4]+ molecular orbitals
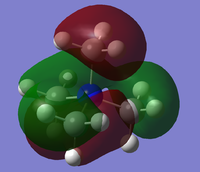
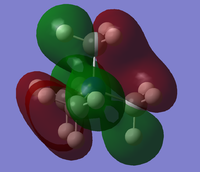
From the population analysis checkpoint file for [N(CH3)4]+, the occupied molecular orbitals were visualised as surfaces. Five were chosen for further analysis:
 |
 |
 |
 |
 |
7 is a highly bonding MO with a single nodal plane. A nitrogen p atomic orbital interacts strongly with two s-like methyl fragment orbitals in each of its lobes, giving two large volumes of electron density, each of which fully covers one methyl group and partially covers another, with through-space interactions between each of the two groups.
12 is also a bonding MO with a similar pattern to 7; however, there are now additional nodal planes through the methyl groups suggesting we are seeing the interaction between a nitrogen p orbital and p-type methyl fragment orbitals.. Thus, two pairs of methyl groups have through-space interactions as in 7, but two groups also have opposite-phase electron density covering their H atoms.
15 is a slightly anti-bonding MO with two nodal parabolic surfaces, each running though two carbon atoms and the central atom. Between these two surfaces, four hydrogen atoms distributed in a square around the central atom are all connected by a ring-shaped volume of electron density for weak through-space interactions. Each of the pairs of carbon atoms and remaining four hydrogen atoms on either side are connected by strong through-space interactions. This suggests the interaction of adjacent p-type fragment orbitals with no contribution from the central nitrogen atom.
18 is a highly anti-bonding MO with nodal planes through several σv symmetry planes. Each methyl group has a carbon p orbital mixed with the hydrogen orbitals, but in anti-bonding arrangement with all adjacent orbitals. There are through-space interactions between two hydrogen atoms on each methyl group. There is no electron density around the central nitrogen atom.
20 is an anti-bonding MO with a nodal plane though one methyl group, a parabolic nodal surface through the central atom, and a further parabolic surface through the other three methyl groups. These nodes separate areas of electron density which are composed of through-space interactions between methyl groups. This shows several aligned p-type orbitals, of which only two, those of the nitrogen and the first methyl group, are arranged for a totally bonding interaction.
Charge distributions and bond contributions
| Molecule | X contribution to X-C bond | X atom charge | C atom charge | H atom charge |
|---|---|---|---|---|
| [N(CH3)4]+ | 66.35% | -0.295 | -0.483 | 0.269 |
| [P(CH3)4]+ | 40.43% | 1.667 | -1.060 | 0.298 |
| [S(CH3)3]+ | 51.33% | 0.917 | -0.840 | 0.297 & 0.279 |
The positive charge is not placed on any single atom but is distributed across the whole molecule in a pattern which can be related to the electronegativities of the central atoms, which go in the order N (3.04) > S (2.58) > C (2.55) > P (2.19). Nitrogen has such high electronegativity that it carries a slight negative charge. Sulfur, having an electronegativity very close to that of carbon, comes close to having the formal +1 charge. Phosphorous has a lower electronegativity than carbon so has a higher-than-expected positive charge.
The charges on the carbon atoms are all negative but vary in magnitude proportionally with the charges on the central atoms. The charges on the hydrogen atoms vary slightly, but are all roughly +0.3. In trimethylsulfonium, the 'up' H atoms have slightly more charge than the 'down' atoms.
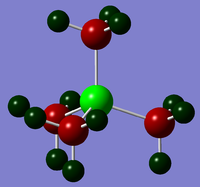
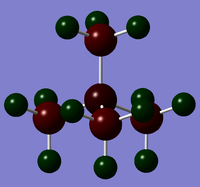 |
 |
 |
Bond contributions follow the same trend as electronegativity; however, the fact that the elements are on different rows can also be considered. Nitrogen's valence orbitals will be similar in size to those in carbon, thus mixing of the two is more facile and nitrogen can provide a larger orbital contribution to bonding than either phosphorous or sulfur, which have larger orbitals and less overlap with carbon's orbitals.
The typical depiction of [NR4]+ has the formal positive charge on the nitrogen atom, as in the arrow pushing mechanism the lone pair on :NR3 donates to form a dative bond, leaving the nitrogen with 'one fewer' electron. The charge distribution as calculated above shows this not to be the case: in reality, the nitrogen has a slight negative charge, and the positive charge is distributed about the adjacent alkyl hydrogen atoms.
Influence of functional groups
| Molecule | N atom charge |
|---|---|
| [N(CH3)4]+ | -0.295 |
| [N(CH3)3(CH2OH)]+ | -0.322 |
| [N(CH3)3(CH2CN)]+ | -0.289 |
The electron-donating effect of the alcohol group and the electron-withdrawing effect of the nitrile group are reflected in the differing charges on the central N atom in [N(CH3)3(CH2OH)]+ and [N(CH3)3(CH2CN)]+. The alcohol group has a much stronger effect. With the addition of these functional groups, symmetry is broken thus the methyl group atoms no longer have equal charges, thus have not been included on the table. Charge distributions from a range of -1 to 1 can be seen below:
 |
 |
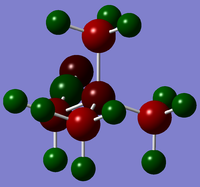 |
The HOMO and LUMO of these three cations were visualised:
| Molecule | HOMO | LUMO | Energy difference |
|---|---|---|---|
| [N(CH3)4]+ |  |
 |
0.44632 |
| [N(CH3)3(CH2OH)]+ |  |
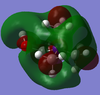 |
0.36304 |
| [N(CH3)3(CH2CN)]+ | 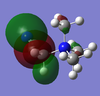 |
 |
0.31864 |
In the HOMOs, the addition of a functional group draws electron density away from the central atom and to the group; this is especially pronounced for electron-withdrawing nitrile. The LUMOs remain similar in shape, but become more diffuse with addition of a functional group. The HOMO energy is increased by addition of a functional group, but the HOMO-LUMO energy gap is lessened. This, together with the larger LUMOs, suggest that the cations with functional groups are more susceptible to reductive reactions. As the HOMO is distributed mostly around the functional group, oxidative and substitution reactions are more likely to happen here.
