Rep:Mod:testosterone420
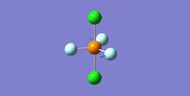
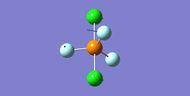
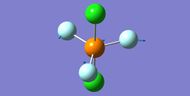
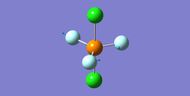
NH3
Interactive structure of NH3 |
Properties
| Molecule | NH3 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(D,P) |
| Final energy (au) | -56.55776 |
| RMS gradient | 0.00000485 |
| Point group | C3V |
| N-H bond length (Å) | 1.02 ± 0.01 |
| H-N-H bond angle | 105.7° ± 1° |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Vibrational Modes
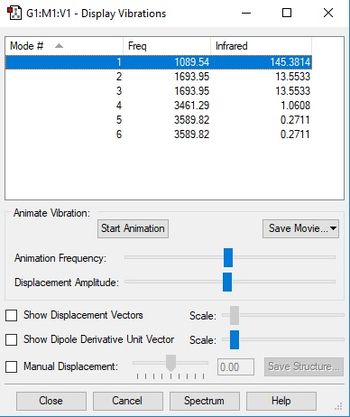
| Wavenumber (cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
|---|---|---|---|---|---|---|
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity (arbitrary units) | 145 | 14 | 14 | 1 | 0 | 0 |
| Image | 
|

|

|
|

|

|
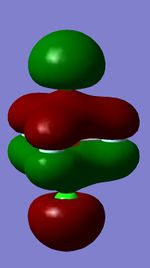
Using the 3N-6 rule, the expected number of vibrational modes is 3 x 4 - 6 = 6 modes. The modes at 1694 cm-1 are degenerate, as well as the modes at 3590 cm-1. The bending modes are the ones at 1090 cm-1 and 1694 cm-1, and the stretching modes are the ones at 3461 cm-1 and 3590 cm-1. The modes that are highly symmetric are the ones at 1090 cm-1 and 3461 cm-1. In an experimental spectrum of gaseous ammonia, the expected number of bands would be 4, because 4 modes are actually pairs of degenerate modes. In reality, it would be very difficult to make out the degenerate vibrational modes at 3461 cm-1 and 3590 cm-1 on a spectrum, because their intensity is so low.
Charge Distribution
Considering the electronegativity of nitrogen is higher than that of hydrogen, the expected charge distribution would be a negative charge on nitrogen and a positive charge on the hydrogens. The calculated charge distribution is on the image below.

N2
Interactive structure of N2 |
Properties
| Molecule | N2 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(D,P) |
| Final energy (au) | -109.52412 |
| RMS gradient | 0.00000060 |
| Point group | D∞h |
| N-N bond length (Å) | 1.11 ± 0.01 |
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Vibrational Modes
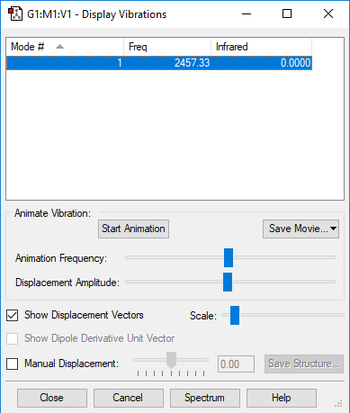
| Wavenumber (cm-1) | 2457 |
|---|---|
| Symmetry | SGG |
| Intensity (arbitrary units) | 0 |
| Image | 
|
Using the 3N-5 rule, the expected number of vibrational modes is 3 x 2 - 5 = 1 mode, which is at 2457 cm-1. It is a stretching mode and since it is symmetric, there is no change in dipole, and it is not IR active. In an experimental spectrum of nitrogen gas, the expected number of bands would be 0, because because there are no modes with a change in dipole.
Charge Distribution
Since a nitrogen molecule consists of 2 nitrogen atoms with the same electronegativity, the overall charge is 0 and the charge on each atom is also 0. The calculated charge distribution is on the image below.
H2
Interactive structure of H2 |
Properties
| Molecule | H2 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(D,P) |
| Final energy (au) | -1.17853 |
| RMS gradient | 0.00000023 |
| Point group | D∞h |
| H-H bond length (Å) | 0.74 ± 0.01 |
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000001 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Vibrational Modes
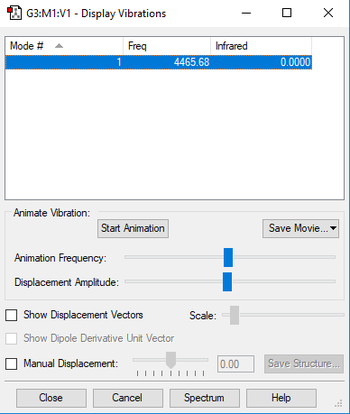
| Wavenumber (cm-1) | 4466 |
|---|---|
| Symmetry | SGG |
| Intensity (arbitrary units) | 0 |
| Image | 
|
Using the 3N-5 rule, the expected number of vibrational modes is 3 x 2 - 5 = 1 mode, which is at 4466 cm-1. It is a stretching mode and since it is symmetric, there is no change in dipole, and it is not IR active. In an experimental spectrum of hydrogen gas, the expected number of bands would be 0, because because there are no modes with a change in dipole.
Charge Distribution
Since a hydrogen molecule consists of 2 hydrogen atoms with the same electronegativity, the overall charge is 0 and the charge on each atom is also 0. The calculated charge distribution is on the image below.

Mono-Metallic Transition Metal Complex Incorporating H2
There is a complex that coordinates H2, where the bond length between the hydrogens is different than in hydrogen gas. The complex is Chloro-(dihydrogen-H,H')-hydrido-tris(triphenylphosphine)-osmium, with the identifier CEFCAS. The H-H bond distance in the complex is 1.482 Å.
The reason for the increased bond length is the that the d orbitals of the osmium atom overlap with the bonding and antibonding orbitals of the two hydrogens, increasing the antibonding character in the H-H bond, weakening it and elongating it. Eventually, this H-H bond gets cleaved, leaving 2 hydride ligands.[1] Also, when calculating the bond lengths with software, some approximations have to be made, which changes the result. Experimentally obtained values may also be affected by external factors.
Interactive structure of the metal complex |
Energy of Formation of Ammonia
The formation of ammonia from hydrogen and nitrogen gas goes as follows:
N2 + 3H2 -> 2NH3
The energy change of this reaction can be found by calculating the energies of each reactant and products, then subtracting the energy of the reactants from the energy of the products. The energies of the reactants and products have been determined in the previus sections.
- E(NH3) = -56.55776 au
- 2 * E(NH3) = -113.11553 au
- E(N2) = -109.52412 au
- E(H2) = -1.17853 au
- 3 * E(H2) = -3.53561 au
- ΔE = 2 * E(NH3) - [E(N2) + 3 * E(H2)]= -113.11553 - (-109.52412 -3.53561) = -0.0557 au
In kJ/mol that value is approximately -146.8. Since ΔE is negative, ammonia must be more stable than the gaseous reactants.
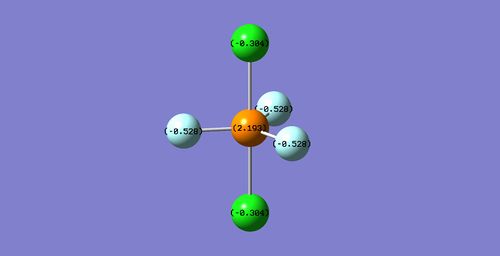
PF3Cl2
Interactive structure of PF3Cl2 |
Properties
| Molecule | PF3Cl2 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(D,P) |
| Final energy (au) | -1561.34405 |
| RMS gradient | 0.00000295 |
| Point group | D3H |
| P-Cl bond length (Å) | 2.10 ± 0.01 |
| P-F bond length (Å) | 1.58 ± 0.01 |
| F-P-F bond angle | 180° |
| F-P-Cl bond angle | 90° |
| Cl-P-Cl bond angle | 120° |
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000003 0.000300 YES Maximum Displacement 0.000046 0.001800 YES RMS Displacement 0.000014 0.001200 YES
Vibrational Modes
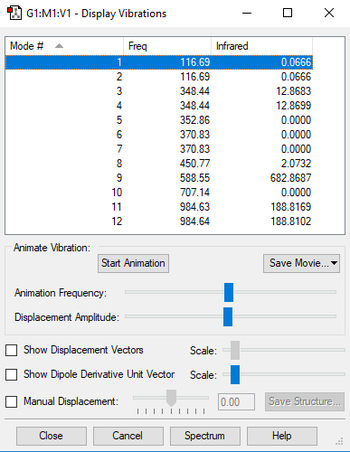
1)Note: for the modes at 353 cm-1 and 371 cm-1, Gaussian was unable to determine symmetry.
Using the 3N-6 rule, the expected number of vibrational modes is 3 x 6 - 6 = 12 modes. There are degenerate pairs of vibrational modes at 116 cm-1, 349cm-1, 371 cm-1, and 985 cm-1. The vibrational modes are the first 7 (up to 371 cm-1), excluding the mode at 353 cm-1. The rest of them are stretching modes. The modes that are highly symmetric are the ones at 353 cm-1 and 707 cm-1. In an experimental spectrum of PF3Cl2, the expected number of bands would be 5, because 6 modes are actually pairs of degenerate modes and 4 modes are IR inactive. In reality, it would be very difficult to make out the degenerate vibrational modes at 117 cm-1 and 451 cm-1 on a spectrum, because their intensity is so low.
Charge Distribution
Considering the electronegativity of fluorine is the highest, chlorine the second highest, and phosphorus the lowest, the expected charge distribution would be a high negative charge on the fluorine atoms, a weaker negative charge on the chlorine atoms, and a positive charge on the phosphorus atom. The calculated charge distribution is on the image below.
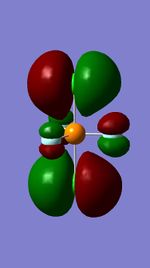
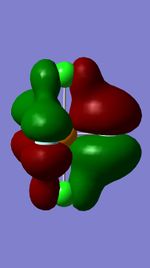
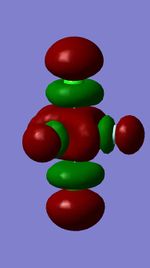
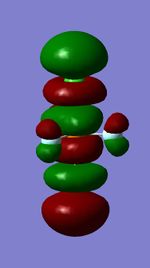
Molecular Orbitals
Bond Lengths of PCl3F2
Interactive structure of PCl3F2 |
The bond lengths for PCl3F2 are different , because of the equatorial and axial positions are in different environments. The P-F bond length in PCl3F2 is 1.62 Å, compared to 1.58 Å for PF3Cl2. The P-Cl bond length in PCl3F2 is 2.05 Å, compared to 2.10 Å in PF3Cl2. The equatorial P-Cl bond length is shorter than the axial P-Cl bond length, just as the equatorial P-F bond length is shorter than the axial P-F bond length. The reason is that the axial bonds are formed from the overlap with an unhybridized phosphorus 3p orbital, while the equatorial bonds are formed from overlap with sp2 hybridized phosphorus orbitals, which have more s character.
References
- ↑ Angew.Chem.Int.Ed. 2005, 44,7227–7230, DOI: 10.1002/anie.200502297
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
I'm not going to mark you down for this but it is completely inappropriate to write "testosterone420" as the title of your page in work that is handed in to be marked. You should only write things relevant to the subject at hand in any piece of academic work.
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, most answers are correct. However there are only 2 visible peaks in the spectra of NH3, due to the low intensity of the other 2 peaks. (See infrared column in vibrations table.)
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - especially good discussion of the charges well done! You could have improved by adding more detail on the MOs - for example the LUMO is antibonding (as you stated) but does not affect the P-X bonding as it is unoccupied.
Independence 0.5/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
YES - you did an extra calculation on PCl3F2 and discussed your results. It would have been good for you to include a link to the log file of the independent calculation.
Do some deeper analysis on your results so far