Rep:Mod:stephen
Introduction to Conformer Searches in Avogadro
When attempting to ascertain the lowest energy conformer/rotamer of a complex molecule the total number of candidates becomes very large very quickly (The aldol product in the second year molecular modelling workshop for example has 5831 possible rotamers arising from just 6 rotatable bonds - presently we will discuss this molecule). [Avogadro] has an interesting feature which can bypass having to optimise each individual conformation accessible by Extensions > Molecular Mechanics > Conformer Search which provides the user with four options:
- Systematic: Simply computes through stable conformations, ending at an arbitrary point. This is only really useful for molecules with 2-3 rotatable bonds as with fewer the results will frequently rubber-band back into their original places after optimisation and with more the total number of conformers is prohibitively large.
- Random: Generates a random set of conformations of the size specified by the user and keeps the lowest energy. Will often return the same result as a weighted search for small/non complex molecules.
- Weighted: Identifies conformations likely to be lower in energy in order to improve efficiency.
- Genetic Algorithm: Darwinian algorithm that discards high energy conformations and mutates low energy conformations for a series of rounds to optimise close to a minimum. This may not, however, be the global minimum but merely a local minimum (as indeed is the case for each of the other search methods).
With the exception of systematic (which overloads the CPU), each of these searches may be conducted while autooptimisation is ongoing so that the returned conformer may be optimised immediately.
Further reading
2nd Year Modelling Crib Sheet (Avogadro)
[All energies are calculated with the MMFF94s force field with the conjugate gradients algorithm. There is a rather interesting bug with avogadro (report here) where dragging hydrogens 'out' of other atoms to bond them causes a runtime error, so if any students manage to cause this advise them to sue Build > Add Hydrogens instead]
Cyclohexane:
Chair: -14.909kJ/mol
Twist boat: 9.91757kJ/mol
[using Build > Invert Chirality actually works on the twist boat form - this may come in handy to explain to students that it is in fact chiral]
Tetrahydropyran
Chair: 15.6315kJ/mol
Twist boat: 41.8728kJ/mol
Butanes
RR = SS = 40.7827kJ/mol (Br and Cl app, methyls gauche)
SR = RS = 43.0077kJ/mol (Hydrogens, Methyls and Halogens app)
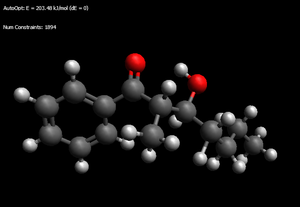
Aldol Product
[It is likely that there are lower energies but these are the lowest I have found]
SR = RS = 173.165kJ/mol (hydrogen bond between carbonyl and hydroxyl, three gauche interactions along the alkyl chain)
SS = 173.927kJ/mol (again hydrogen bond, two gauche interactions in side chain)
RR = 178.658kJ/mol (no hydrogen bond and three gauche interactions - 193.272 for H bond and only two, should give some idea of how much the two effects are relatively worth)
Ciculenes
[5]Circulene =
[7]Circulene = 683.981kJ/mol (planar, Avogadro does not appear to be able to create a puckered conformation)
[8]Circulene = 322.344kJ/mol (planar)
cis Decalins
Chair-chair = 34.2392kJ/mol
Di-twist boat (up/up) = 70.0503kJ/mol
Chair-twist boat = 61.0438kJ/mol
Di-twist boat (up/down) = 75.819kJ/mol
cis-Cortisone = 121.772kJ/mol
trans-Cortisone = 119.184kJ/mol
Cuprate addition species
Methyl up = 66.1288kJ/mol
Methyl down = 69.0429kJ/mol
Grignard addition species
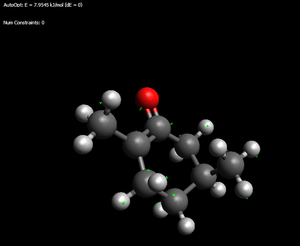
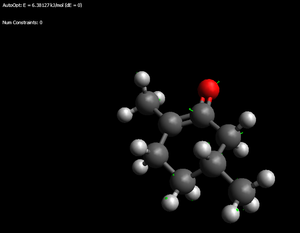
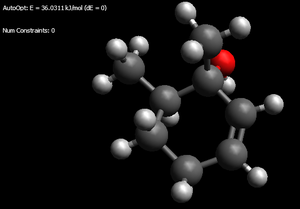
iso-Menthone enols
Thermodynamic enol: 32.1787kJ/mol (the '>' of the isopropyl is pointing at the hydroxyl)
Kinetic enol: 38.4377kJ.mol (again > towards the OH as well as a possible favourable interation - 2.208angstrom - between alkene hydrogen and hydroxyl hydrogen)
Menthones
R,S Menthone = 61.0112kJ/mol
R,R Menthone = 62.7216kJ/mol
(is this close enough to 0.7kJ/mol to be acceptable?)
Deprotonated enols (deprotonating clockwise from bottom left):
280.198kJ/mol
353.588kJ/mol
163.817kJ/mol (green colouring giving the answer away?...)
302.642kJ/mol
