Rep:Mod:shine11
Inorganic Computational Chemistry
Optimisation
BH3
3-21G
The log file for the optimisation is Media:JS_BH3_OPT3.LOG
All bond distances are reported to two decimal places, bond angles are reported to one and the dipole moment is reported to two. This is a result of the fact that the program is only accurate to this level of precision.
The optimised B-H bond distance is 1.20Ao and the optimum bond angle is 120.0o.
| BH3 | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Final energy/au | -26.4622 |
| Gradient/au | 0.00004507 |
| Dipole moment/D | 0.00 |
| Point group | D3h |
| Time taken/s | 7.0 |
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000352 0.001800 YES
RMS Displacement 0.000230 0.001200 YES
Predicted change in Energy=-4.580958D-08
Optimization completed.
-- Stationary point found.
6-31G(d,p)
The log file for this, more accurate optimisation, is Media:JS_BH3_OPTACCURATE.LOG.
The optimised average bond length is 1.20Ao and the optimised bond angle is 120.0o.
| BH3 | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31g(d,p) |
| Final energy/au | -26.6153 |
| Gradient/au | 0.00000673 |
| Dipole moment/D | 0.00 |
| Point group | Cs |
| Time taken/s | 13.0 |
Item Value Threshold Converged?
Maximum Force 0.000011 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.000056 0.001800 YES
RMS Displacement 0.000036 0.001200 YES
Predicted change in Energy=-9.815639D-10
Optimization completed.
-- Stationary point found.
GaBr3
The optimised bond length is 2.35Ao and the optimised bond angle is 120.0o. This compares reasonably well to a literature vale of 2.38Ao[1].
| GaBr3 | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | LANL2DZ |
| Final energy/au | -41.7008 |
| Gradient/au | 0.00000016 |
| Dipole moment/D | 0.00 |
| Point group | D3h |
| Time taken/s | 52.1 |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
Predicted change in Energy=-1.282692D-12
Optimization completed.
-- Stationary point found.
BBr3
The optimised bond length is 1.93Ao and the optimised bond angle is 120.0o.
| BBr3 | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -64.43644 |
| Gradient/au | 0.00001054 |
| Dipole moment/D | 0.00 |
| Point group | Cs |
| Time taken/s | 20.7 |
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000011 0.000300 YES
Maximum Displacement 0.000167 0.001800 YES
RMS Displacement 0.000082 0.001200 YES
Predicted change in Energy=-2.420020D-09
Optimization completed.
-- Stationary point found.
Bond Length Comparison
| Bond Length/Ao | |
|---|---|
| BH3 | 1.19 |
| BBr3 | 1.93 |
| GaBr3 | 2.35 |
Changing the ligand from H to Br significantly increases the bond length. The reasons for this are numerous. Firstly, Br has 3 more full s and p orbitals and 1 more full d orbital than H. It is also has three lone pairs, one of which can donate into the empty B p orbital, since the B is sp2 hybridised. This actually shortens the bond length. The other lone pair in the p orbital can cause repulsion which lengthens the bond. Changing the central atom from B to Ga also increases the bond length. First, Ga has two extra s and p orbitals, and a full d orbital, which will repel the orbitals on Br. Second, the fact that Ga is less electronegative than B will lengthen the bond, since the Ga will attract the electrons in the bond less strongly.
Defining a chemical bond is not as trivial as the conventional use of a straight line to indicate covalent bond would make it appear. Typically, it is a potential energy minimum of a system, existing as a result of an attractive electrostatic interaction over a distance that is generally between 1.4-3Ao. The strength of these interaction can vary greatly, from the covalent bonding in N2, to the hydrogen bonds in water. However, many of these interactions exist in molecules, and simply using distance to determine the existence or non-existence of a bond, as Gaussian does, can lead to important interactions not being detected. In molecular orbital theory, any stabilised orbital is considered to be a bonding orbital. Clearly, this is a more accurate method for the determination of bond formation than just using distance, although every single interaction cannot, of course be considered to be a bond. A combination of both methods must be used.
Frequency Analysis
BH3
This log file for this process is Media:JS_BH3_FREQ3.LOG.
Low frequencies --- -0.9580 -0.9214 -0.0053 5.3695 11.5406 11.5803 Low frequencies --- 1162.9950 1213.1817 1213.1844
| BH3 | |
|---|---|
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Final energy/au | -26.4622 |
| Gradient/au | 0.00008904 |
| Dipole moment/D | 0.00 |
| Point group | Cs |
| Time taken/s | 26.0 |
Item Value Threshold Converged? Maximum Force 0.000221 0.000450 YES RMS Force 0.000106 0.000300 YES Maximum Displacement 0.000724 0.001800 YES RMS Displacement 0.000445 0.001200 YES Predicted change in Energy=-1.667569D-07 Optimization completed. -- Stationary point found.
Vibration Visualisation
The optimised average bond length is 1.20Ao and the optimised bond angle is 120.0o. Force constant related to freq. Gaussian bases bonds on distance
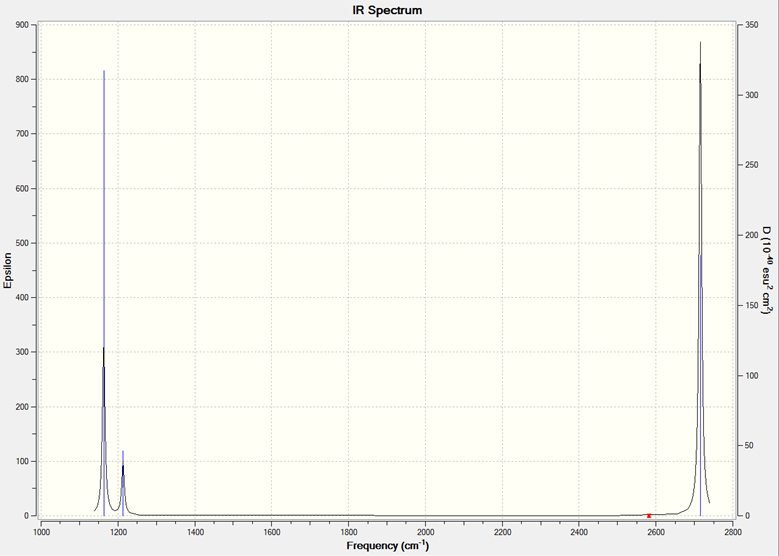
The actual IR spectrum is shown below.
GaBr3
-2.3539 -2.3538 -1.3645 -0.0026 0.0044 0.0276
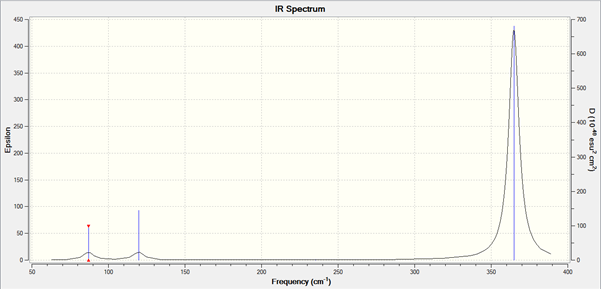
The lowest "real" normal mode is 86.9748cm-1.
The IR spectrum is below.
Vibration Comparison
The frequencies of BH3 are much greater than for GaBr3, for the same vibrational mode. The main reasons for this are that, first, the frequency is inversely proportional to the reduced mass. Since the reduced mass of GaBr3 is obviously much greater, its frequencies are reduced. Second, frequency is proportional to the force constant of the bond. The Ga-Br bond is longer than the B-H bond, suggesting that it is a weaker bond. Therefore, its frequencies are again reduced.
Interestingly, bends 2 and 3 are at higher frequency than bend 1 for BH3, but are at lower frequency than bend 1 for GaBr3. Apart from this, the spectra a reasonably similar, apart from the fact that the GaBr3 A2 bend has a lower intensity. Bends 1,2 and 3 are at similar frequencies, since they are bends. Stretches 4,5 and 6 are also at similar frequencies, because they are stretches. The intensity of stretch 4 is zero, since this stretch causes no change in dipole moment of the molecule. There are only three peaks seen due to the degeneracy of the E' vibrations.
This frequency analysis gives eigenvalues that represent the second derivatives of the energy diagram. Therefore, if they are positive, the energy is a minimum. The low frequencies are to do with the idea that each molecule has 3N-6 degrees of freedom, specifically the "-6", meaning the motions of the center of mass of the molecule. Clearly, the same method and basis set must be used for both pieces of analysis, in order for the results to be compared fairly.
BH3 MO Analysis
The line "int=ultrafine scf=conver=9" was used to improve the values for the low frequencies.
The log file for the optimisation is Media:JS_BH3_OPT.LOG
Item Value Threshold Converged?
Maximum Force 0.000003 0.000015 YES
RMS Force 0.000002 0.000010 YES
Maximum Displacement 0.000013 0.000060 YES
RMS Displacement 0.000008 0.000040 YES
Predicted change in Energy=-6.228076D-11
Optimization completed.
-- Stationary point found
The log file for the frequency is Media:JS BH3 FREQ2.LOG.
-11.2361 -11.2272 -5.6980 0.0003 0.0322 0.4463
The D-space for the population analysis is here.
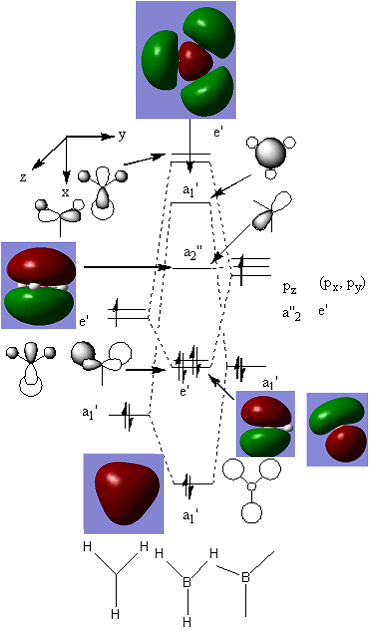
This is the MO diagram with all modeled occupied orbitals and the modeled LUMO.
Comparing the generated orbitals to what MO theory predicted, most of the orbitals appear to be reasonable models of the actual MO, considering the speed of the method. However, the first e' orbital is difficult to view with this program. Also, the rest of the orbitals lack the symmetry that is expected. From this, it also seems that MO theory is useful, for qualitative predictions.
NH3 NBO Analysis
The log file for the optimisation is Media:JS NH3 OPT.LOG.
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629715D-09
Optimization completed.
-- Stationary point found.
The log file for the frequency is Media:JS NH3 FREQ2.LOG.
-9.3920 -8.3783 -6.3678 -0.0017 -0.0017 -0.0009
This is the D-space for the population analysis.
This is the charge distribution.
The charge range was -1.125 to 1.125
| Atom | NBO charge |
|---|---|
| N | -1.125 |
| H | +0.375 |
NH3BH3 Energy Minimum
The log file for the optimisation is Media:JS NH3BH3 OPT3.LOG.
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000026 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-2.956782D-10
Optimization completed.
-- Stationary point found.
The log file for the frequency is Media:JS NH3BH3 FREQ.LOG.
The low frequencies are -6.4842 -4.4175 -0.0011 -0.0009 -0.0008 0.8459.
Energy comparison
E(NH3)= -56.55776872a.u. E(BH3)= -26.6153237196a.u. E(NH3BH3)= -83.22468907 a.u.
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE=(-83.22468907)-[(-26.6153237196)+(-56.55776872)]
= -0.0516 a.u.
= -0.0516*2625.5 KJ/mol
= -135 KJ/mol
Schlenk Equilibrium
Considering the grignard reagent MeMgCl, the following species were analysed, excluding Me2Mg and MgCl2.
(1) MeMgCl
Optimisation
A mixture of basis sets were used. 6-31G(d,p) was used for the C and H atoms, while LanL2DZ was used for the Mg and Cl atoms. The log file for this optimisation is Media:JS Mg mix opt.log.
The optimised bond length for Mg-Cl is 2.26Ao and for Mg-Me is 2.07Ao. The optimised bond angle is 180.0o.
| MeMgCl | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -55.769 |
| Gradient/au | 0.00000838 |
| Dipole moment/D | 3.13 |
| Point group | C1 |
| Time taken/s | 68 |
Item Value Threshold Converged?
Maximum Force 0.000012 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000396 0.001800 YES
RMS Displacement 0.000145 0.001200 YES
Predicted change in Energy=-2.482616D-09
Optimization completed.
-- Stationary point found.
Frequency Analysis
This log file for this process is Media:JS MEMGCL FREQ.LOG.
Low frequencies --- -17.8458 -3.0113 -0.0001 -0.0001 0.0002 4.3589
| MeMgCl | |
|---|---|
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -55.769 |
| Gradient/au | 0.00000843 |
| Dipole moment/D | 3.13 |
| Point group | C1 |
| Time taken/s | 38 |
Item Value Threshold Converged?
Maximum Force 0.000029 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.000472 0.001800 YES
RMS Displacement 0.000199 0.001200 YES
Predicted change in Energy=-3.505925D-09
Optimization completed.
-- Stationary point found.
Vibrations
(2) Me2Mg2Cl2L2, L = Et2O
Optimisation
Here is the optimisation file Media:JS MG2 MIX ET2O OPT3.LOG
| MG2, L = ET2O | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -579.0039 |
| Gradient/au | 0.00000342 |
| Dipole moment/D | 9.51 |
| Point group | C1 |
| Time taken | 1h 43min |
Item Value Threshold Converged?
Maximum Force 0.000010 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000537 0.001800 YES
RMS Displacement 0.000115 0.001200 YES
Predicted change in Energy=-2.800254D-09
Optimization completed.
-- Stationary point found.
Frequency
This is the frequency analysis file Media:JS Mg2 Et2O FREQ.log
| Mg2, L=Et2O | |
|---|---|
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -579.004 |
| Gradient/au | 0.00000343 |
| Dipole moment/D | 9.50 |
| Point group | C1 |
| Time taken | 1h 8min 40s |
Low frequencies --- -4.2354 0.0005 0.0007 0.0008 4.7788 8.7652
Vibrations
Unfortunately, there were 120 generated vibrational modes for these molecules, making it difficult to analyse each mode individually. Therefore, only the key modes are shown, meaning the ones that involve the Mg and Cl atoms.
A key point to note, that is evidenced by the following spectra, as well as the population analysis, is that the diagram at the beginning of this section is only a very rough estimate of where the electrons actually lie. Specifically, there appears to be a high amount of electron density between the two Cl atoms. The term "bond" is not used here, due to the vagueness of its definition, discussed above. The modes 1, 5, 7 and 9 seem to show this. An advantage of using this program is that the symmetric stretches of Cl-Cl can still be viewed, despite the fact that they are not IR active, since they do not cause a change in the dipole of the molecule. These all have A1 symmetry. Due to the fact that the frequency of the vibration is inversely proportional to the reduced mass, the Cl-Cl stretches and bends are at a low frequency. In addition, there seems to be a so called "bond" between the Mg and Cl atoms, shown by modes 2, 3, 4 and 6. Furthermore, mode 9 suggests the presence of a bond between the Mg atoms.
Comparing these modes to those for MeMgCl, they are very different, in terms of their frequencies, suggesting that the solvent molecule is interacting with the molecule quite profoundly. For instance, mode number 3 for MeMgCl has a frequency 250cm-1 greater than mode number 3 for the one above, which is the same mode. This indicates that the Mg has become electron deficient. This is the only mode that can be compared fairly, based on the fact that the ligand is also affecting the reduced mass, as well as the force constant. Therefore, it is not a clear indication of the presence of electron density.
Molecular orbitals
This is the D-space for the population analysis.
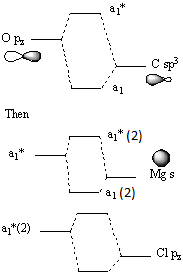
Molecular orbital 45 is the px orbital on Cl, overlapping with the Mg-O bond. Considering the C-O bond, it formed by the s orbital on the Mg mixing with an O orbital, since they are both have a1 symmetry. The O orbital consists of an sp3 orbital on the C, mixing with the pz orbital on the O.
The vast majority of these modes indicate that there is a reasonably large amount of electron density between the Cl atoms, that is, it could be said that there is a "bond" between them. Interestingly, energy level 45 shows that, in addition, the O can overlap with the central Mg and Cl atoms as well. Also, energy level 47 shows a high level of electron density between all of the central Mg and Cl atoms, as does the LUMO. This explains why the IR spectrum indicates the existence of Mg-Mg and Mg-Cl "bonds". Furthermore, the HOMO involves the central atoms, as well as the methyl group. The majority of the electron density is on the Me, since it is the C that is electronegative and acting as a nucleophile, shown in the NBO analysis below.
Natural Bond Orbital Analysis
This image of charge distribution is from -1.375 to 1.375, with green indicating positive charge.
| Atom | Mean NBO charge |
|---|---|
| Mg | +1.280 |
| C | -1.374 |
| Cl | -0.654 |
| O | -0.66 |
The C is the most electronegative atom, which can then act as a nucleophile in C-C bond forming reactions, as expected. The Mg is electropositive, stabilized by the electronegative O, which comes from the solvent.
Looking at the log file gives a significant amount of information about the central atoms.
1. (1.97330) BD ( 1)Mg 1 -Cl 3
( 5.76%) 0.2399*Mg 1 s( 23.62%)p 3.23( 76.38%)
0.4858 0.0130 -0.5089 -0.0444 -0.1705
-0.0157 0.6871 0.0388
( 94.24%) 0.9708*Cl 3 s( 30.28%)p 2.30( 69.72%)
0.5503 0.0007 0.7286 0.0012 0.1048
0.0005 -0.3942 0.0030
2. (1.97364) BD ( 1)Mg 1 -Cl 4
( 5.93%) 0.2435*Mg 1 s( 24.67%)p 3.05( 75.33%)
0.4965 0.0111 -0.4100 -0.0316 -0.2557
-0.0247 -0.7184 -0.0467
( 94.07%) 0.9699*Cl 4 s( 30.58%)p 2.27( 69.42%)
0.5530 0.0005 0.6706 0.0013 0.1856
0.0007 0.4582 -0.0029
4. (1.97265) BD ( 1)Mg 2 -Cl 3
( 5.89%) 0.2427*Mg 2 s( 24.73%)p 3.04( 75.27%)
-0.4972 -0.0105 -0.4259 -0.0316 0.1065
0.0117 -0.7460 -0.0471
( 94.11%) 0.9701*Cl 3 s( 29.52%)p 2.39( 70.48%)
-0.5434 -0.0001 0.6809 0.0014 -0.0389
-0.0002 0.4895 -0.0033
5. (1.97283) BD ( 1)Mg 2 -Cl 4
( 5.74%) 0.2395*Mg 2 s( 23.67%)p 3.22( 76.33%)
-0.4863 -0.0150 -0.5228 -0.0335 0.2379
0.0185 0.6563 0.0343
( 94.26%) 0.9709*Cl 4 s( 29.49%)p 2.39( 70.51%)
-0.5431 -0.0001 0.7394 0.0011 -0.1085
0.0008 -0.3827 0.0028
Considering the Mg-Cl bonds, it is clearly a relatively accurate approximation to say that the Mg atoms are sp3 hybridised. This means that their ideal bond angle should be somewhere around 109o. The fact that their average bond angles are actually 98.6o and 122.9o suggests that there is a significant amount of ring strain. This is obviously greater in the Mg3 molecule, which could be one reason why it is the least stable molecule. Looking at the Cl atoms, which constitute almost 95% of the bonds, they are all almost exactly 3:7 s:p orbitals. These would, again, prefer an angle of around 115o. The fact that the internal angles are almost 90o, again, suggests that a large amount of ring strain is present.
No Mg-Mg or Cl-Cl bonds are officially detected by Gaussian, due to the previously stated fact that it detects the existence of bonds based on the distances between the atoms. It is clear from the analysis that they can be said to exist, however.
(2) Me2Mg2Cl2L2, L = THF
Optimisation
This is the D-space for the optimisation.
| MG2, L = THF | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -576.581 |
| Gradient/au | 0.00017625 |
| Dipole moment/D | 10.41 |
| Point group | C1 |
| Time taken | 4h 7min 46s |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.001290 0.001800 YES
RMS Displacement 0.000296 0.001200 YES
Predicted change in Energy=-4.797915D-09
Optimization completed.
-- Stationary point found.
Frequency
Here is the D-space for the frequency analysis.
| MG2, L = THF | |
|---|---|
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -576.581 |
| Gradient/au | 0.00000763 |
| Dipole moment/D | 10.45 |
| Point group | C1 |
| Time taken | 54min 20s |
Low frequencies --- -11.6009 -0.0004 -0.0004 -0.0003 2.8476 7.2269
Molecular orbitals
This is the D-space for the population analysis.
(3) Me2Mg2Cl2L2, L = THF
Optimisation
This is the D-space for the optimisation.
| MG2, L = THF | |
|---|---|
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -576.555 |
| Gradient/au | 0.00025064 |
| Dipole moment/D | 13.28 |
| Point group | C1 |
| Time taken | 4h 26min 9s |
Item Value Threshold Converged? Maximum Force 0.002152 0.000450 YES RMS Force 0.000281 0.000300 YES Maximum Displacement 0.454039 0.001800 YES RMS Displacement 0.110593 0.001200 YES Predicted change in Energy=-5.882623D-04
Frequency
This is the D-space for the frequency analysis.
| MG2, L = THF | |
|---|---|
| File type | .log |
| Calculation type | FREQ |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy/au | -576.559 |
| Gradient/au | 0.00001015 |
| Dipole moment/D | 11.41 |
| Point group | C1 |
| Time taken | 4h 40min 59s |
Molecular orbitals
This is the log file Media:JS Mg3 THF NBO.log and this is the checkpoint file Media:JS Mg3 THF NBO FINAL.fchk for the population analysis.
Energy Comparison
| Energy of Molecule/a.u. | |
|---|---|
| Mg2, L = Et2O | -579.0039 |
| Mg2, L = THF | -576.581 |
| Mg3, L = THF | -576.555 |
Looking at these values, it is clear that considering their absolute values in KJ/mol would be pointless, due to their magnitude. However, when the structures are viewed, they all look sensible. Therefore, their energies can be compared to each other.
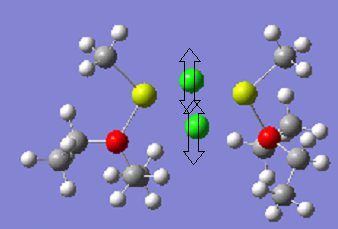
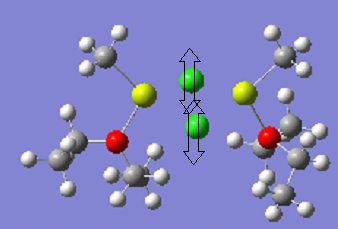
Interestingly, Mg2 with L=THF is more stable than Mg3 with L=THF. This suggests that, overall, the intramolecular forces between the protons on the THF ligands and the methyl groups are attractive, and are lowering the energy of the molecule. Viewing the molecules below shows that the Mg3 molecule, shown , is trying to maximise this interaction. The , is already close to the optimum distance.
The shortest distance between the protons is 2.33Ao in the Mg molecule, but 2.67Ao in Mg2. However, Mg2 has many more of these interactions than Mg2. Presumably, the molecule with Et2O ligands is the most stable due to the lack of ring strain. In addition, it is easier for molecule to form the H-H interactions.
MO Comparison
The orbitals of each molecule have almost identical geometry, therefore analysing the energy of these orbitals are key to understanding their chemistry. Based on this, the Mg2, L=Et2O, is the best electrophile, from the fact that it has the lowest lying LUMO. Mg3, L=THF, is the best nucleophile, with its highest energy HOMO.
Conclusion
This particular grignard reagent is a reasonably reactive molecule. The reasons for this are numerous, and are shown clearly by the population analysis, with the Mg3 molecule being the slightly better nucleophile. This analysis also shows that both solvents stabilize the molecules, with Et2O dong a slightly better job, shown by their relative energies. In addition, the population analysis shows explicitly what exactly is happening, in terms of bonding between the central atoms. Using molecular orbital theory, it was possible to explain what exactly was mixing in the majority of the key orbitals. As well as this, the vibrational analysis was in agreement with the population analysis and used to reinforce the data. The log file from the population analysis, in particular, was useful in analysing the position of the electron density between the central atoms. The two difficulties encountered were that, first, the Mg3, L=Et2, molecule was not successfully generated. Second, the issue that Gaussian is unable to label the interactions between Mg-Mg and Cl-Cl as bonds.
References
- ↑ H. Ulrich and G Heyne, Physikal. Chem., 1941, B 49, 284