Rep:Mod:seaunicorn12
A note on accuracy
Values in summary tables are quoted to the full number of decimal places as given by Gaussian. Any calculated values are given to fewer decimal places to account for errors and inaccuracy in calculations.
Mod 2: Week 1, Day 1
Part 2: BH3 Molecule optimisation 3-21
The BH3 bond lengths were set to 1.5Å as the starting point.
Optimisation file
File:Vm1110 BH3 OPT.LOG
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 3-21G
Charge: 0
Spin: Singlet
E(RB3LYP): -26.46226338 au
RMS Gradient Norm: 0.00020672 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 39.0 seconds.
The symmetry assigned to the molecule by Gaussian corresponds to the actual symmetry of the molecule, no symmetry restrictions required for further steps.
Item table
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Part 7: BH3 Molecule optimisation 6-31G(d,p)
Optimisation file
File:Vm1110 BH3 OPT2.LOG
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -26.61532252 au
RMS Gradient Norm: 0.00021672 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 31.0 seconds.
The B-H bond length in the BH3 molecule had decreased from the first optimisation from 1.193 to 1.1914 Å. This is a change of 0.02 ± 0.01 Å.
Item table
Item Value Threshold Converged?
Maximum Force 0.000433 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001702 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.189019D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1914 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1914 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1914 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Mod 2: Week 1, Day 2
Part 1: TlBr3 Molecule optimization LanL2DZ
The point group was restricted to D3H on very tight (0.0001) tolerance.
DOI:10042/21611
File:Vm1110 TlBr opt lanL2DZ.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: LANL2DZ
Charge: 0
Spin: Singlet
E(RB3LYP): -91.21812851 au
RMS Gradient Norm: 0.00000090 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 19.2 seconds.
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084147D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Optimised bond length: 2.65 Å (literature: 2.5 Å[1])
Optimised Br-Tl-Br bond angle: 120°
The reported bond length from the optimization is 2.65 ± 0.01 Å which is a little different from the literature value of 6.5 Å[1]. The difference in value could be attributed to the bond length being measured in a different molecule. Also, slight disparities between theoretical calculation and experimentally determined values.
Part 2: BBr3 Molecule optimization [Gen: B(6-31G(d,p); Br(LanL2DZ)]
The structure of the BBr3 molecule was based on the optimized BH3 molecule with H atoms replaced with Br atoms.
<bt>
DOI:10042/21622
File:Vm1110 Bbr3 opt gen2.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -64.43645296 au
RMS Gradient Norm: 0.00000383 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 10.9 seconds.
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000046 0.001800 YES
RMS Displacement 0.000030 0.001200 YES
Predicted change in Energy=-5.266345D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Optimised bond length: 1.934 Å
Optimised Br-B-Br bond angle: 120°
Part 3: Comparing BH3, BBr3 and TlBr3 bond lengths
| BH3 | BBr3 | TlBr3 |
| 1.19 | 1.93 | 2.65 |
Bond length discussion
The central atom-ligand bond increases in the order B-H < B-Br < Tl-Br.
From the values for the B-H and B-Br bond lengths, certain information can be deduced regarding the effect of changing the ligand has on bond length. The bond length increases from 1.19 Å to 1.93 Å which is a 0.74 ± 0.01 Å increase. H is a much smaller atom then Br (smaller covalent and van der Waal radii) which means that when it bonds, it bonds much closer to the central atom, hence a shorter bond length. Br and H are similar in that they both require 1 electron to fulfill their octet and doublet respectively. They are also both quite electronegative, Br more so than H, which means that their orbitals are not very diffuse and the bond lengths are shorter than would be for less electronegative atoms.
Comparing the B-Br bond length with the Tl-Br bond length gives the insight on how changing the central atom has on the bond length between central atom and ligand. Boron and Thallium are both in group 13 so they have the same valence electron structure and require the same number of electrons to complete their octet. Boron is slightly more electronegative than Thallium and as such has less diffuse orbitals. However, as Thallium is in group 6 (vs group 2 for Boron) it has many more orbitals and as such this is the largest contributing factor to it's larger size (rather than electronegativity). The increased size of the atom increases the bond length in this case B-Br and Tl-Br bond lengths are 1.93 Å and 2.65 Å respectively and the increase in bond length is 0.72 ± 0.01 Å.
What is a bond?
Gaussview does not always draw bonds between atoms within a structure even if the bonds exist. Gaussview draws in bonds below a pre-defined value for the particular bond in question. For example, in the optimisation of BH3, a bond length of 1.4 Å is not drawn as a visible bond but a bond of 1.2 Å is. This does not mean that there is no bond, Gaussview assigns bonds according to the distance between atoms rather than bonding or non bonding character of the molecular orbitals. <bt> A bond can be defined in one way as the existence of electrons in molecular orbitals of lower energy than the atomic orbitals, with a largely bonding character. In that way if there are equally as many electrons in antibonding orbitals as there are in bonding orbitals, a bond will not form because the molecule is not stabilized. In addition, any electrons residing in non-bonding orbitals do not contribute to the overall bond.
Mod 2: Day 3
Part 2: BH3 Molecule frequency
File:VM1110 BH3 FREQ.LOG
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -26.61532252 au
RMS Gradient Norm: 0.00021669 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 8.0 seconds.
Low frequencies --- -70.3431 -69.5542 -69.5540 -0.0055 0.0695 0.1595 Low frequencies --- 1161.4019 1212.1084 1212.1111
The low frequencies do not fall into the ±15 range however there are no negative vibrational modes which indicates that the molecule was in fact optimized to a minimum. The large range for the low frequencies may be in part due to the basis set used or the symmetry of the molecule. As it is a very small molecule it is possible to have lower accuracy and therefore larger errors.
Part 3: BH3 Molecule vibrations
IR spectrum
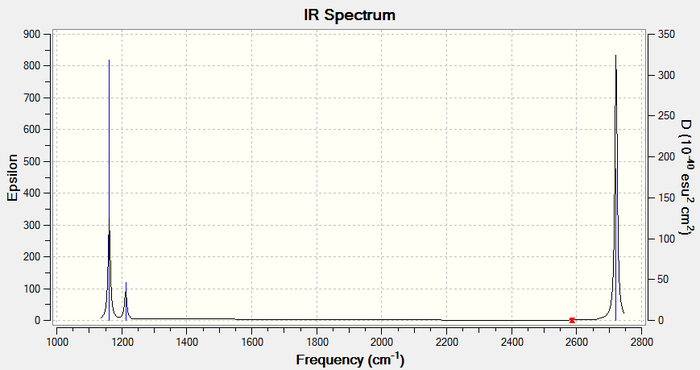
Only 3 peaks are visible in the IR spectrum. This is because vibrations 5 and 6 and, 2 and 3 have the same frequency and therefore show up as a single peak. In addition, vibration 4 has an intensity of 0, hence it has no peak on the spectrum.
Part 4: TlBr3 Molecule frequency
DOI:10042/22056
File:Vm1110 TlBr opt lanL2DZ freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: LANL2DZ
Charge: 0
Spin: Singlet
E(RB3LYP): -91.21812851 au
RMS Gradient Norm: 0.00000088 au
Dipole Moment: 0.0000 Debye
Point Group: D3H
Job cpu time: 22.0 seconds.
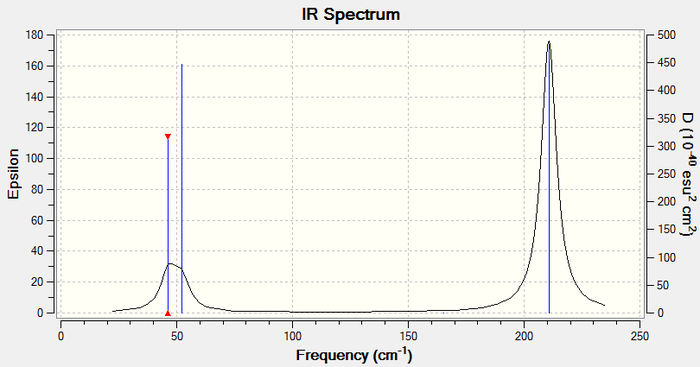
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
Lowest frequency mode: 46.4289 (E')
Ir Spectrum
Vibrational frequency comparison
| Number | Boron | Thallium | ||
|---|---|---|---|---|
| Frequency | Symmetry | Frequency | Symmetry | |
| 1 | 1161.40 | A2" | 46.4289 | E' |
| 2 | 1212.11 | E' | 46.4292 | E' |
| 3 | 1212.11 | E' | 52.1449 | A2" |
| 4 | 2587.81 | A1' | 165.2685 | A1' |
| 5 | 2721.52 | E' | 210.6948 | E' |
| 6 | 2721.52 | E' | 210.6948 | E' |
By comparison it is possible to see that the frequencies for TlBr3 are much lower than for BH3. This shows that the Tl-Br bonds are much weaker than B-H bonds. In addition to the vast difference in frequencies, there has also been a reordering of the modes. Where as in BH3 the lowest frequency mode was A2" in TlBr3 the lowest frequency mode is E' symmetry and the A2" mode is of the third lowest frequency.
The two spectra are similar in the manner that the both have two peaks at a lower frequency and one large peak at the highest frequency. The peaks on the thallium spectrum are much broader and far less intense. Also there is more of an overlap of the two peaks in the lower frequency region leading to a more spread double pointed peak rather than two distinct peaks. The A1' and E' vibrations lie at a higher frequency for both spectra because they are of a higher energy than the A2" and E' vibrations.
Frequency calculations
Frequency calculations are run using the same basis set as the optimizations because these determine the symmetry of the molecule and the way in which the energy is measured. The lowest energy version of a molecule in one basis set might not be the lowest energy conformation in another basis set. Hence for the frequency analysis to work the energy must be at a minimum and the basis set must be the same.
The frequency analysis gives information regarding the vibrations of the molecule at its lowest energy state. This information can be used to find which vibrations are IR active and hence create a theoretical IR spectrum.
The 6 low frequencies are the motion of the molecule's centre of mass. When these values are low it means the molecule has been fully optimized and the frequency analysis is not affected by the motion of the overall molecule in measuring the molecule's vibrations.
Part 5: BH3 Molecular Orbitals
DOI:10042/22065
File:Vm1110 BH3 MOs.fchk
File Type: .fch
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -26.53393782 au
RMS Gradient Norm: 0.00000000 au
Dipole Moment: 0.0000 Debye
Job cpu time: 0.9 seconds.
| Molecular orbital Diagram | Calculated molecular orbitals |
|---|---|
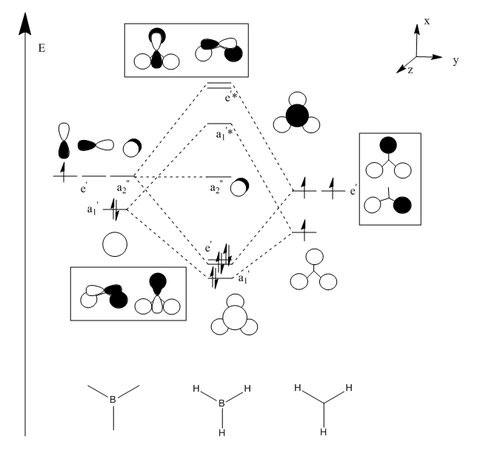
|
 LUMO (a2") |
 (e') | |
 (e') | |
 (a1') |
Discussion
The LCAO orbitals and the calculated orbitals are all quite similar in appearance, the calculated orbitals more combined to give one overall orbital rather rather than distinct overlapped orbitals. However they are still quite similar. This shows that in this case LCAO theory works well to define what is observed through calculation.
Mod 2: Day 4
Part 1: NBO Analysis
NH3 optimisation
DOI:10042/22066
File:VM1110 NH3 OPT 631 GDP 2.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -56.55776856 au
RMS Gradient Norm: 0.00000888 au
Dipole Moment: 1.8464 Debye
Point Group: C1
Job cpu time: 8.7 seconds.
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000088 0.001800 YES
RMS Displacement 0.000056 0.001200 YES
Predicted change in Energy=-1.759459D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7414 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7478 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3 frequency calculation
DOI:10042/22067
File:VM1110 NH3 freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -56.55776856 au
RMS Gradient Norm: 0.00000888 au
Dipole Moment: 1.8464 Debye
Point Group: C1
Job cpu time: 17.0 seconds.
Low frequencies --- -30.7764 -0.0020 -0.0018 -0.0012 20.3142 28.2484 Low frequencies --- 1089.5557 1694.1237 1694.1868
NH3 population analysis
File:VM1110 NH3 pop.log
File Type: .log
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -56.55776856 au
RMS Gradient Norm: au
Dipole Moment: 1.8464 Debye
Point Group: C1
Job cpu time: 15.8 seconds.
NH3 NBO analysis
| NH3 charge distribution (-1.0 to +1.0) | |
|---|---|
| |
| N Charge | H Charge |
| -1.125 | 0.375 |
Part 2: Association energy
NH3BH3 optimisation
DOI:10042/
File:VM1110 NH3BH3 opt 631 gdp 2.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -83.22468730 au
RMS Gradient Norm: 0.00005971 au
Dipole Moment: 5.5649 Debye
Point Group: C1
Job cpu time: 16.8 seconds.
Item Value Threshold Converged?
Maximum Force 0.000122 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000662 0.001800 YES
RMS Displacement 0.000295 0.001200 YES
Predicted change in Energy=-1.639776D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.21 -DE/DX = -0.0001 !
! R2 R(2,8) 1.2101 -DE/DX = -0.0001 !
! R3 R(3,8) 1.2101 -DE/DX = -0.0001 !
! R4 R(4,7) 1.0186 -DE/DX = -0.0001 !
! R5 R(5,7) 1.0186 -DE/DX = -0.0001 !
! R6 R(6,7) 1.0186 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(4,7,5) 107.8722 -DE/DX = 0.0 !
! A2 A(4,7,6) 107.8744 -DE/DX = 0.0 !
! A3 A(4,7,8) 111.0188 -DE/DX = 0.0 !
! A4 A(5,7,6) 107.8673 -DE/DX = 0.0 !
! A5 A(5,7,8) 111.0335 -DE/DX = 0.0 !
! A6 A(6,7,8) 111.0296 -DE/DX = 0.0 !
! A7 A(1,8,2) 113.8814 -DE/DX = 0.0 !
! A8 A(1,8,3) 113.879 -DE/DX = 0.0 !
! A9 A(1,8,7) 104.5977 -DE/DX = 0.0 !
! A10 A(2,8,3) 113.8671 -DE/DX = 0.0 !
! A11 A(2,8,7) 104.5945 -DE/DX = 0.0 !
! A12 A(3,8,7) 104.5926 -DE/DX = 0.0 !
! D1 D(4,7,8,1) 179.9734 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -60.0193 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 59.9696 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -60.027 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 59.9803 -DE/DX = 0.0 !
! D6 D(5,7,8,3) 179.9692 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 59.9736 -DE/DX = 0.0 !
! D8 D(6,7,8,2) 179.9809 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -60.0302 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3BH3 frequency
DOI:10042/
File:VM1110 NH3BH3 freq 2.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Charge: 0
Spin: Singlet
E(RB3LYP): -83.22468730 au
RMS Gradient Norm: 0.00005971 au
Dipole Moment: 5.5649 Debye
Point Group: C1
Job cpu time: 1 minute 8.9 seconds.
Low frequencies --- -13.5525 -0.0010 -0.0008 -0.0006 16.5718 28.9370 Low frequencies --- 265.0672 632.1225 638.0187
Energy comparison
| Molecule | Energy |
|---|---|
| NH3 | -56.55776856 au |
| BH3 | -26.61532252 au |
| NH3BH3 | -83.22468730 au |
| ΔE | -0.052 au (-135.47kJ/mol) |
Dissociation energy: -135.47 kJ/mol
Mod 2: Week 2, AlCl2Br dimer isomers
Part 1: AlCl2Br dimer optimisations
Isomer 1
File:Vm1110 AlCl4Br2 isomer 1 opt gen 3.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.40630797 au
RMS Gradient Norm: 0.00000327 au
Dipole Moment: 0.0012 Debye
Point Group: C2V
Job cpu time: 28.4 seconds.
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000324 0.001800 YES
RMS Displacement 0.000097 0.001200 YES
Predicted change in Energy=-1.843622D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 2.0934 -DE/DX = 0.0 !
! R2 R(2,8) 2.0934 -DE/DX = 0.0 !
! R3 R(3,7) 2.0934 -DE/DX = 0.0 !
! R4 R(4,7) 2.0934 -DE/DX = 0.0 !
! R5 R(5,7) 2.4893 -DE/DX = 0.0 !
! R6 R(5,8) 2.4893 -DE/DX = 0.0 !
! R7 R(6,7) 2.4893 -DE/DX = 0.0 !
! R8 R(6,8) 2.4893 -DE/DX = 0.0 !
! A1 A(7,5,8) 88.2675 -DE/DX = 0.0 !
! A2 A(7,6,8) 88.2675 -DE/DX = 0.0 !
! A3 A(3,7,4) 121.7613 -DE/DX = 0.0 !
! A4 A(3,7,5) 109.8017 -DE/DX = 0.0 !
! A5 A(3,7,6) 109.8017 -DE/DX = 0.0 !
! A6 A(4,7,5) 109.813 -DE/DX = 0.0 !
! A7 A(4,7,6) 109.813 -DE/DX = 0.0 !
! A8 A(5,7,6) 91.7325 -DE/DX = 0.0 !
! A9 A(1,8,2) 121.7613 -DE/DX = 0.0 !
! A10 A(1,8,5) 109.8017 -DE/DX = 0.0 !
! A11 A(1,8,6) 109.8017 -DE/DX = 0.0 !
! A12 A(2,8,5) 109.813 -DE/DX = 0.0 !
! A13 A(2,8,6) 109.813 -DE/DX = 0.0 !
! A14 A(5,8,6) 91.7325 -DE/DX = 0.0 !
! D1 D(8,5,7,3) -111.7781 -DE/DX = 0.0 !
! D2 D(8,5,7,4) 111.8043 -DE/DX = 0.0 !
! D3 D(8,5,7,6) 0.006 -DE/DX = 0.0 !
! D4 D(7,5,8,1) 111.7781 -DE/DX = 0.0 !
! D5 D(7,5,8,2) -111.8043 -DE/DX = 0.0 !
! D6 D(7,5,8,6) -0.006 -DE/DX = 0.0 !
! D7 D(8,6,7,3) 111.7781 -DE/DX = 0.0 !
! D8 D(8,6,7,4) -111.8043 -DE/DX = 0.0 !
! D9 D(8,6,7,5) -0.006 -DE/DX = 0.0 !
! D10 D(7,6,8,1) -111.7781 -DE/DX = 0.0 !
! D11 D(7,6,8,2) 111.8043 -DE/DX = 0.0 !
! D12 D(7,6,8,5) 0.006 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Isomer 2
File:Vm1110 AlCl4Br2 isomer 3 opt gen 3.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41632880 au
RMS Gradient Norm: 0.00002607 au
Dipole Moment: 0.1920 Debye
Point Group: CS
Job cpu time: 54.9 seconds.
Item Value Threshold Converged?
Maximum Force 0.000083 0.000450 YES
RMS Force 0.000031 0.000300 YES
Maximum Displacement 0.000571 0.001800 YES
RMS Displacement 0.000264 0.001200 YES
Predicted change in Energy=-6.679677D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,3) 2.0926 -DE/DX = 0.0 !
! R2 R(2,3) 2.0926 -DE/DX = 0.0 !
! R3 R(3,6) 2.2958 -DE/DX = 0.0 !
! R4 R(3,7) 2.2958 -DE/DX = 0.0 !
! R5 R(4,5) 2.2765 -DE/DX = 0.0 !
! R6 R(4,6) 2.3006 -DE/DX = 0.0 !
! R7 R(4,7) 2.3006 -DE/DX = 0.0 !
! R8 R(4,8) 2.2765 -DE/DX = 0.0 !
! A1 A(1,3,2) 121.7655 -DE/DX = 0.0 !
! A2 A(1,3,6) 110.0697 -DE/DX = 0.0 !
! A3 A(1,3,7) 110.0697 -DE/DX = 0.0 !
! A4 A(2,3,6) 110.0696 -DE/DX = 0.0 !
! A5 A(2,3,7) 110.0696 -DE/DX = 0.0 !
! A6 A(6,3,7) 90.3043 -DE/DX = -0.0001 !
! A7 A(5,4,6) 110.2008 -DE/DX = 0.0 !
! A8 A(5,4,7) 110.2008 -DE/DX = 0.0 !
! A9 A(5,4,8) 121.5012 -DE/DX = 0.0 !
! A10 A(6,4,7) 90.0628 -DE/DX = -0.0001 !
! A11 A(6,4,8) 110.2008 -DE/DX = 0.0 !
! A12 A(7,4,8) 110.2008 -DE/DX = 0.0 !
! A13 A(3,6,4) 89.8164 -DE/DX = 0.0001 !
! A14 A(3,7,4) 89.8164 -DE/DX = 0.0001 !
! D1 D(1,3,6,4) -111.5487 -DE/DX = 0.0 !
! D2 D(2,3,6,4) 111.5488 -DE/DX = 0.0 !
! D3 D(7,3,6,4) 0.0001 -DE/DX = 0.0 !
! D4 D(1,3,7,4) 111.5487 -DE/DX = 0.0 !
! D5 D(2,3,7,4) -111.5488 -DE/DX = 0.0 !
! D6 D(6,3,7,4) -0.0001 -DE/DX = 0.0 !
! D7 D(5,4,6,3) 111.6137 -DE/DX = 0.0 !
! D8 D(7,4,6,3) -0.0001 -DE/DX = 0.0 !
! D9 D(8,4,6,3) -111.6138 -DE/DX = 0.0 !
! D10 D(5,4,7,3) -111.6137 -DE/DX = 0.0 !
! D11 D(6,4,7,3) 0.0001 -DE/DX = 0.0 !
! D12 D(8,4,7,3) 111.6138 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Isomer 3
File:Vm1110 AlCl4Br2 isomer 4 opt gen 2.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41626675 au
RMS Gradient Norm: 0.00002016 au
Dipole Moment: 0.1669 Debye
Point Group: CS
Job cpu time: 52.6 seconds.
Item Value Threshold Converged?
Maximum Force 0.000049 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000512 0.001800 YES
RMS Displacement 0.000174 0.001200 YES
Predicted change in Energy=-2.899010D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.0939 -DE/DX = 0.0 !
! R2 R(2,5) 2.2981 -DE/DX = 0.0 !
! R3 R(2,6) 2.2981 -DE/DX = 0.0 !
! R4 R(2,7) 2.2747 -DE/DX = 0.0 !
! R5 R(3,4) 2.2747 -DE/DX = 0.0 !
! R6 R(3,5) 2.2981 -DE/DX = 0.0 !
! R7 R(3,6) 2.2981 -DE/DX = 0.0 !
! R8 R(3,8) 2.0937 -DE/DX = 0.0 !
! A1 A(1,2,5) 109.8477 -DE/DX = 0.0 !
! A2 A(1,2,6) 109.8477 -DE/DX = 0.0 !
! A3 A(1,2,7) 121.4998 -DE/DX = 0.0 !
! A4 A(5,2,6) 90.1702 -DE/DX = 0.0 !
! A5 A(5,2,7) 110.5144 -DE/DX = 0.0 !
! A6 A(6,2,7) 110.5144 -DE/DX = 0.0 !
! A7 A(4,3,5) 110.5164 -DE/DX = 0.0 !
! A8 A(4,3,6) 110.5164 -DE/DX = 0.0 !
! A9 A(4,3,8) 121.4893 -DE/DX = 0.0 !
! A10 A(5,3,6) 90.1678 -DE/DX = 0.0 !
! A11 A(5,3,8) 109.8534 -DE/DX = 0.0 !
! A12 A(6,3,8) 109.8534 -DE/DX = 0.0 !
! A13 A(2,5,3) 89.829 -DE/DX = 0.0 !
! A14 A(2,6,3) 89.829 -DE/DX = 0.0 !
! D1 D(1,2,5,3) 110.7482 -DE/DX = 0.0 !
! D2 D(6,2,5,3) -0.4771 -DE/DX = 0.0 !
! D3 D(7,2,5,3) -112.519 -DE/DX = 0.0 !
! D4 D(1,2,6,3) -110.7482 -DE/DX = 0.0 !
! D5 D(5,2,6,3) 0.4771 -DE/DX = 0.0 !
! D6 D(7,2,6,3) 112.519 -DE/DX = 0.0 !
! D7 D(4,3,5,2) 112.5206 -DE/DX = 0.0 !
! D8 D(6,3,5,2) 0.4771 -DE/DX = 0.0 !
! D9 D(8,3,5,2) -110.7542 -DE/DX = 0.0 !
! D10 D(4,3,6,2) -112.5206 -DE/DX = 0.0 !
! D11 D(5,3,6,2) -0.4771 -DE/DX = 0.0 !
! D12 D(8,3,6,2) 110.7542 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Isomer 4

File:Vm1110 AlCl4Br2 isomer 5 opt gen 2.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41631606 au
RMS Gradient Norm: 0.00001798 au
Dipole Moment: 0.0059 Debye
Point Group: CS
Job cpu time: 54.8 seconds.
Item Value Threshold Converged?
Maximum Force 0.000046 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000619 0.001800 YES
RMS Displacement 0.000158 0.001200 YES
Predicted change in Energy=-2.701528D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,4) 2.2984 -DE/DX = 0.0 !
! R2 R(1,5) 2.2984 -DE/DX = 0.0 !
! R3 R(1,7) 2.0938 -DE/DX = 0.0 !
! R4 R(1,8) 2.2746 -DE/DX = 0.0 !
! R5 R(2,3) 2.2746 -DE/DX = 0.0 !
! R6 R(2,4) 2.2985 -DE/DX = 0.0 !
! R7 R(2,5) 2.2985 -DE/DX = 0.0 !
! R8 R(2,6) 2.0937 -DE/DX = 0.0 !
! A1 A(4,1,5) 90.1599 -DE/DX = 0.0 !
! A2 A(4,1,7) 109.8678 -DE/DX = 0.0 !
! A3 A(4,1,8) 110.5187 -DE/DX = 0.0 !
! A4 A(5,1,7) 109.8678 -DE/DX = 0.0 !
! A5 A(5,1,8) 110.5187 -DE/DX = 0.0 !
! A6 A(7,1,8) 121.4683 -DE/DX = 0.0 !
! A7 A(3,2,4) 110.4933 -DE/DX = 0.0 !
! A8 A(3,2,5) 110.4933 -DE/DX = 0.0 !
! A9 A(3,2,6) 121.5473 -DE/DX = 0.0 !
! A10 A(4,2,5) 90.1547 -DE/DX = 0.0 !
! A11 A(4,2,6) 109.8433 -DE/DX = 0.0 !
! A12 A(5,2,6) 109.8433 -DE/DX = 0.0 !
! A13 A(1,4,2) 89.8427 -DE/DX = 0.0 !
! A14 A(1,5,2) 89.8427 -DE/DX = 0.0 !
! D1 D(5,1,4,2) -0.0017 -DE/DX = 0.0 !
! D2 D(7,1,4,2) -111.2475 -DE/DX = 0.0 !
! D3 D(8,1,4,2) 112.0414 -DE/DX = 0.0 !
! D4 D(4,1,5,2) 0.0017 -DE/DX = 0.0 !
! D5 D(7,1,5,2) 111.2475 -DE/DX = 0.0 !
! D6 D(8,1,5,2) -112.0414 -DE/DX = 0.0 !
! D7 D(3,2,4,1) 112.0113 -DE/DX = 0.0 !
! D8 D(5,2,4,1) 0.0017 -DE/DX = 0.0 !
! D9 D(6,2,4,1) -111.2123 -DE/DX = 0.0 !
! D10 D(3,2,5,1) -112.0113 -DE/DX = 0.0 !
! D11 D(4,2,5,1) -0.0017 -DE/DX = 0.0 !
! D12 D(6,2,5,1) 111.2123 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
AlCl2Br dimer energy differences
| Isomer | Energy (au) | ΔE (au) | ΔE (kJ/mol) |
|---|---|---|---|
| 1 | -2352.40630797 | 0.0100 | 26.26 |
| 2 | -2352.41632880 | 0 | 0 |
| 3 | -2352.41626675 | 0.0001 | 0.26 |
| 4 | -2352.41631606 | 0.00001 | 0.02 |
Discussion
The isomer of lowest energy is isomer 2 and the isomer of highest energy is isomer 1. Isomer 1 has 2 bridging Br atoms whereas isomers 2, 3 and, 4 have bridging Cl atoms. Hence the dimer is most stable with bridging Cl atoms rather than Br. Isomer 2 has both the terminal Br atoms on one side. Isomers 3 and 4 have the terminal Br atoms on opposite ends in cis and trans conformations respectively. The trans isomer is slightly lower in energy than the cis isomer and both are relatively similar in energy to isomer 2. Hence the molecule is most stable when the Br atoms are terminal and on the same end of the dimer.
Momomer optimization
File:Vm1110 AlCl2Br monomer opt gen 3.log
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -1176.19013667 au
RMS Gradient Norm: 0.00013439 au
Dipole Moment: 0.1194 Debye
Point Group: C2V
Job cpu time: 9.5 seconds.
Item Value Threshold Converged?
Maximum Force 0.000181 0.000450 YES
RMS Force 0.000118 0.000300 YES
Maximum Displacement 0.001031 0.001800 YES
RMS Displacement 0.000577 0.001200 YES
Predicted change in Energy=-2.606668D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.27 -DE/DX = -0.0002 !
! R2 R(1,3) 2.0883 -DE/DX = 0.0002 !
! R3 R(1,4) 2.0883 -DE/DX = 0.0002 !
! A1 A(2,1,3) 120.0373 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0373 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9254 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Dimer dissociation energy
| Molecule | Energy (au) | Energy (kJ/mol) |
|---|---|---|
| monomer | -1176.19013667 | - |
| dimer | -2352.41632880 | - |
| ΔE | -1176.226 | -3.088 106 |
Discussion
Isomer 2 of the dimers is much more stable than the monomer on its own. This is shown by the very large dissociation energy.
Part 2: AlCl2Br dimer frequencies
Isomer 1
DOI:10042/
File:Vm1110 AlCl4Br2 isomer 1 freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.40630797 au
RMS Gradient Norm: 0.00000327 au
Dipole Moment: 0.0012 Debye
Point Group: C2V
Job cpu time: 2minutes 1.7 seconds.
Low frequencies --- -5.1814 -4.9633 -3.1838 0.0030 0.0038 0.0039 Low frequencies --- 14.8246 63.2887 86.0876

|
Isomer 2
DOI:10042/
File:Vm1110 AlCl4Br2 isomer 3 freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41632880 au
RMS Gradient Norm: 0.00002607 au
Dipole Moment: 0.1920 Debye
Point Group: CS
Job cpu time: 3 minutes 46.9 seconds.
Low frequencies --- -3.5964 -2.5820 -1.4168 -0.0020 0.0011 0.0027 Low frequencies --- 17.7720 51.0000 72.1620

|
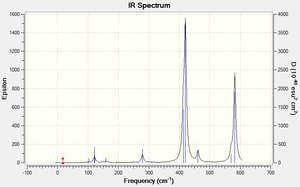
Isomer 3
DOI:10042/
File:Vm1110 AlCl4Br2 isomer 4 freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41626675 au
RMS Gradient Norm: 0.00002016 au
Dipole Moment: 0.1669 Debye
Point Group: CS
Job cpu time: 3 minutes 11.8 seconds.
Low frequencies --- -4.0622 -2.3951 -0.0038 -0.0035 -0.0022 1.0273 Low frequencies --- 17.1580 50.9238 78.5379
|
Isomer 4
DOI:10042/
File:Vm1110 AlCl4Br2 isomer 5 freq.log
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41631606 au
RMS Gradient Norm: 0.00001798 au
Dipole Moment: 0.0059 Debye
Point Group: CS
Job cpu time: 3 minutes 40.1 seconds.
Low frequencies --- -0.0013 0.0020 0.0021 2.1254 2.5567 4.1893 Low frequencies --- 18.1395 49.0727 72.9198
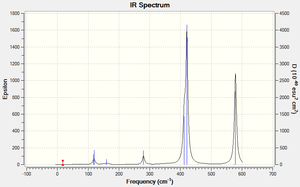
|
IR spectra discussion
More intense bands appear in the spectrum for isomer 1 because more vibrations change the overall dipole of the molecule. In isomers 1 and 2 the Br atoms are close to one another which means that with every vibration there is more electron density moving, leading to more, intense vibrations which show up as a greater number of intense peaks on the IR spectra. Isomer 1 has 18 vibrations and 10 of those are IR inactive. However, of the ones that are IR active, 4 are of a high intensity. The other three isomers have intense IR peaks at approx. 420 which are more intense than any of the isomer 1 peaks but rather than 4, they only have 3 peaks of the same level of intensity. This means that isomer 1 has more vibrations that make a significant change in the overall dipole of the molecule and isomers 3 and 4 have the fewest.
IR discussion
For a vibration to be IR active it needs to create a change in the overall dipole of the molecule. All vibrations that do not create a change are then IR inactive and show an intensity of 0.
Al-Br bond stretch discussion
| Molcule | Vibrations | ||
|---|---|---|---|
| Frequency | Images | Description | |
| Isomer 1 | 196.90 | 0.0 | Al sideways movemenmt in opposite directions, slight Br movement. |
| 241.00 | 99.7720 | Al opposite movement towards and from Br. | |
| 246.75 | 0.0 | Al concerted inward movement. | |
| 341.32 | 160.6510 | Al concerted sideways movement. | |
| 467.22 | 346.5254 | Al opposite movement towards and from Br, Cl outward stretch. | |
| 493.90 | 0.0 | Al inward movement, Cl outward movement, Br fixed. | |
| Isomer 1 | 186.76 | 1.5858 | Al concerted inwards movemenmt, Br movement toward Al. |
| 270.68 | 13.3495 | Al movement outwards in opposite directions, Br moment towards Al. | |
| 418.55 | 309.6868 | Al between Br atoms outward movement, other atoms relatively fixed. | |
| 495.50 | 133.7192 | Al between Br atoms sideways movement, other atoms relatively fixed. | |
| 502.68 | 104.9317 | Al between Cl atoms inward movement, other atoms relatively fixed. | |
Mod 2: Week 2, AlCl2Br lowest energy dimer
Isomer 2 MO calculation
DOI:10042/
File:Vm1110 AlCl4Br2 isomer 3 mo.fchk
File Type: .fchk
Calculation Type: SP
Calculation Method: RB3LYP
Basis Set: Gen
Charge: 0
Spin: Singlet
E(RB3LYP): -2352.41632880 au
RMS Gradient Norm: 0.0000000 au
Dipole Moment: 0.1920 Debye
Point Group: CS
Job cpu time:
MO visualisations
The above table shows a selection of 5 filled non-core molecular orbitals ranging from the most bonding (top) to very antibonding (bottom).