Rep:Mod:ryza0708
Initial Optimisations
BH3 Optimisation (3-21G)
The standard B-H bond lengths after creating a molecule of BH3 were 1.18A. These were manually changed to 1.5A, and then an optimisation carried out, using the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 3-21G
Charge: 0
Spin: singlet
The optimisation log file is linked to here
| heading | heading | |
|---|---|---|
| File Name | bh3-optim | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 3-21G | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.46226338 | a.u. |
| RMS Gradient Norm | 0.00020672 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 20.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
B-H bond distance after optimisation is 1.19A, and the H-B-H bond angle is 120.0 degrees (unchanged).
BH3 Optimisation 6-31G (d,p)
The BH3 molecule was optimised again using the (better) basis set of 6-31G (d,p), as below:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G d, p
Charge: 0
Spin: singlet
The optimisation log file is linked to here
| heading | heading | |
|---|---|---|
| File Name | BH3-631g-dp | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532363 | a.u. |
| RMS Gradient Norm | 0.00000235 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 12.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The B-H bond length is 1.1923, the H-B-H angle is still 120 degrees. As we can see, using the 6-31G basis set, the core non-bonding 1s on Boron are described more accurately, such that the optimisation has resulted in a lowering of the molecules energy than compared to using the 3-21G basis set.
Total energy for the 3-21G optimised structure was -26.46226338 au. Total energy for the 6-31G(d,p) optimised structure was -26.61532363 au. The energy difference is 0.15306025 au.
TlBr3 Optimisation
A molecule of TlBr3 was created with D3H restricted symmetry and very tight tolerance (0.0001). Tl - B bond length was 2.69A. It was optimized as per the below method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: LanL2DZ
Charge: 0
Spin: singlet
Tl - B bond length: 2.65095 Br-Tl-Br bond angle: 120.000
A literature reference for isolated TlBr3 has been difficult to find, likely as TlBr3 exists as a dimer in the gas phase or is, as in the below reference, bonded to solvent. Nonetheless the figure quoted is 2.7035 (B3LYP, experimental 2.5069), which is in good proximity to our calculated figure.
Parlak et al, Erciyes Üniversitesi Fen Bilimleri Enstitüsü Dergisi 27(2): 208-211 (2011)
| heading | heading | |
|---|---|---|
| File Name | log_69143 | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.00000090 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 38.6 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084088D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BBr3 Optimisation
BBr3 was created by replacing H with Br on the 6-31G optimised BH3 structure. It was then optimised as per the below:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: GEN
Charge: 0
Spin: singlet
With "pseduo=read gfinput" entered as an additional keyword. The input file was manually edited to the below before carrying out optimisation:
%chk=\\ic.ac.uk\homes\rb610\3rd Year Lab\BH3-631G-DP.chk # freq b3lyp/gen geom=connectivity gfinput pseudo=read BH3 Optimisation 0 1 B 0.00000000 0.00000000 0.00000000 Br 0.00000000 2.02000000 0.00000000 Br 1.74937146 -1.00999975 0.00000000 Br -1.74937146 -1.00999975 0.00000000 1 2 1.0 3 1.0 4 1.0 2 3 4 B 0 6-31G(d,p) **** Br 0 LanL2DZ **** Br 0 LanL2DZ
| heading | heading | |
|---|---|---|
| File Name | log_69207 | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | Gen | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -64.43645296 | a.u. |
| RMS Gradient Norm | 0.00000382 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 34.8 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027553D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Summary
| Molecule | Bond Length |
|---|---|
| BH3 | 1.19232 |
| TlBr3 | 2.65095 |
| BBr3 | 1.93396 |
To compare the bond lengths of the molecules we have investigated so far, we see that the bond lengths are in the order TlBr3 > BBr3 > BH3.
Br and H both bond covalently to the central boron, but Br has lone pairs that can be donated to the orthogonal p-orbital on boron. This should shorten the bond length through better overlap. In this case, BH3 still has the shorter bond length, as the comparative effect of the Br donation against the smaller size of H is less relevant.
Changing the central ligand from Tl (atomic radius 170pm) to Boron (90pm) decreases the bond length. That is, a larger central atom results in a longer bond, for the same ligands.
Tl and B are similar in that they both covalently bond to the ligands, yet Tl has larger s and p orbitals, resulting in weaker overlap with Br than for B.
In some structures Gaussview does not draw the binds where we expect, however this does not mean that there is no bond, Gaussview creates bonds in a molecule based on distance criteria, that is if there is sufficient orbital interaction between two atoms at that distance, which is set to a pre-defined value. So, there may be orbital interactions beyond this distance, which Gaussview would not class as bonds - even though in reality such a bond may exist.
The definition I would choose for the existence of a bond is: the interaction between two or more atoms of filled, bonding molecular orbitals.
Frequency Analysis
Frequency analysis is used to confirm if the optimisation has successfully created minimised structures.
| heading | heading | |
|---|---|---|
| File Name | ryanbrowne_bh3_freq | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532181 | a.u. |
| RMS Gradient Norm | 0.00027724 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | C3H | |
| Job cpu time: 0 days 0 hours 0 minutes 8.0 seconds. | ||
Total energy for frequency analysis: -26.61532181 au
Total energy for previous optimsation: -26.61532363 au
The frequency analysis log file is linked to here
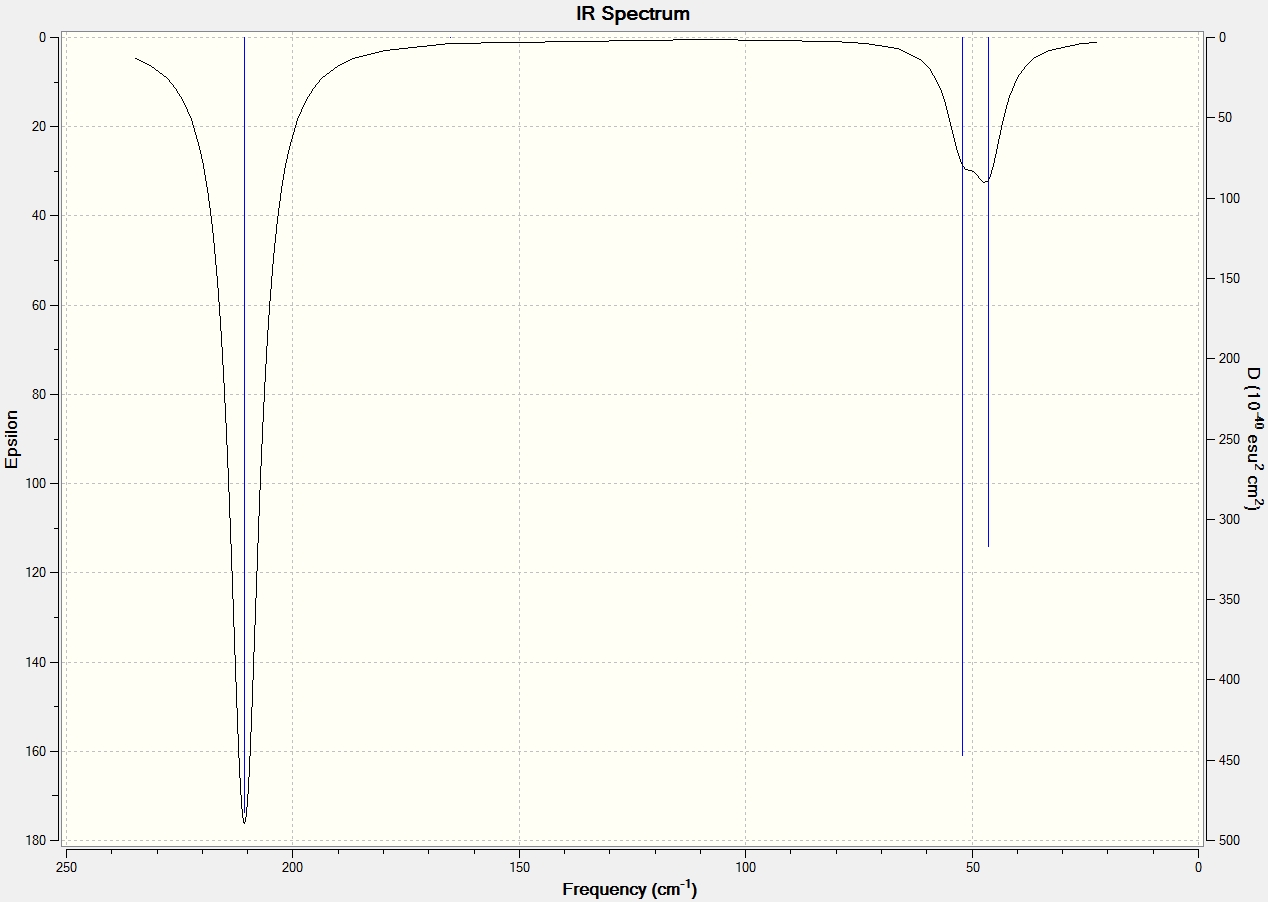
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
The important information to note from the above frequency analysis extract is that the low frequencies top line values fall within the acceptable range of +15cm^-1. This indicates that the molecule has indeed been correctly optimized.
Vibrations
There are six vibrational modes but only three peaks are observed in the spectrum as vibrations 2/3 and 5/6 are degenerate, and hence overlap at the same frequencies, whilst vibration 4 is IR inactive as there is no induced dipole moment, hence the vibration has zero intensity and is not observed in the IR spectrum.
TlBr3 Frequency Analysis
| heading | heading | |
|---|---|---|
| File Name | ryanbrowne_bh3_freq | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.00000088 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 22.9 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
As for BBr3, only three peaks are seen in the IR spectrum as a result of five modes being IR active with two pairs of these modes degenerate.
Summary
| Vibration | BBr3 | TlBr3 |
|---|---|---|
| 1 | 1163.00 (92.5478) | 52.14 (5.8466) |
| 2 | 1213.19 (14.0553) | 46.43 (3.6867) |
| 3 | 1213.19 (14.0589) | 46.43 (3.6867) |
| 4 | 2582.26 (0.0000) | 165.27 (0.0000) |
| 5 | 2715.43 (126.3307) | 210.69 (25.4830) |
| 6 | 2715.43 (126.3211) | 210.69 (25.4797) |
The large difference in the value of the frequencies for BH3 compared to TlBr3 arises as a result of the shorter B-H bond length compared to Tl-Br. Hence to achieve the same vibrational motion, greater energy must be supplied to BBr3. The intensities are lower for TlBr3 as both are less electronegative than B and H respectively, so have more diffuse orbitals. There has been a reordering of the A2" and rocking E' modes in TlBr3. This is as a result of the central atom moving in the A2" and not in the rocking E', and with the greater mass difference between Tl and Br (46au) compared to B and H (4au), this A2" mode is more energetic for TlBr than the E' motion. The spectra are similar in that they each show three peaks. Both molecules have D3H symmetry as they are trigonal planar, and so have six vibrational modes (by 3N-6, 3(4)-6=6). Of these six, only five are IR active (induce a dipole moment), and one is IR inactive. Of the five active modes, two pairs are degenerate in energy and so overlap in frequency. For both spectra, two modes lie fairly closely together, the A2 and E' modes and then the other two modes also lie fairly close together, the A1' and E' modes, but higher in energy. This is because the A2" and E' modes are wagging and scissoring motions, which are more energetic than than the A1' and E' asymmetric, symmetric stretches, as the wagging and scissoring motions cause a change in electron density.
The same method and basis set must be used for both the optimisation and frequency analysis calculations in order to make a fair comparison - using a different method would result in a different potential energy surface, with the energy minimum inequivalent for the optimised molecules. The frequency analysis is carried out so that it can be checked that the molecule has been optimised to an energy minimum. The low frequencies represent the "-6" of the 3N-6.
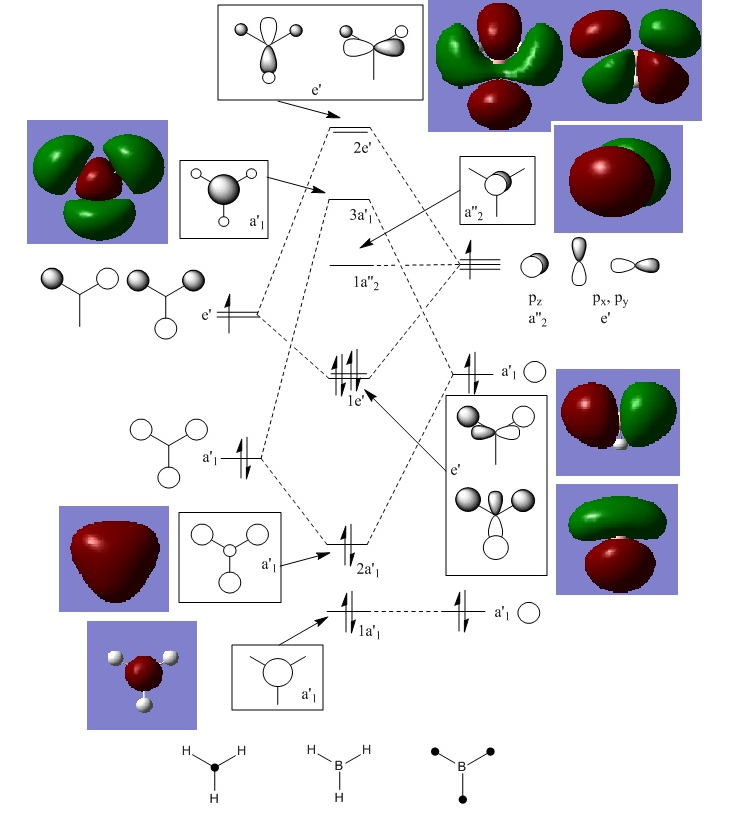
BH3 Molecular Orbitals
Using optimized BH3, a population analysis was run with 6-31G (d, p) basis set, additional keyword "pop=full", and full NBO included. [| D-space link]
There are not any significant differences between the real and LCAO MOs. The LCAO model does not show the merged electron density as is shown in the "real" MOs in Gaussview, but it is clear to see that the merged shapes are created by the combination of similar LCAO phases, and warped or bent away from areas of opposite phase. This suggests that the LCAO model is accurate and useful in qualitative MO theory.
NBO Analysis of NH3
Optimisation
NH3 was optimised as per the below method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: 0
Spin: singlet
The optimisation log file is linked to here
| heading | heading | |
|---|---|---|
| File Name | NH3 optimisation_ 631G | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776856 | a.u. |
| RMS Gradient Norm | 0.00000885 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.8464 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 0 minutes 17.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629727D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7479 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7486 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
N-H Bond length before optmisation: 1.00A
N-H Bond length after optimisation: 1.02A
H-N-H Bond angle before optimisaton: 109.47 degrees
H-N-H Bond angle after optimisation: 105.75 degrees
Frequency Analysis
| heading | heading | |
|---|---|---|
| File Name | NH3 freq | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776856 | a.u. |
| RMS Gradient Norm | 0.00000881 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.8464 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 0 minutes 11.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000077 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.600242D-09
Optimization completed.
-- Stationary point found.
Low frequencies --- -30.6965 -0.0016 0.0009 0.0012 20.2203 28.2940 Low frequencies --- 1089.5540 1694.1243 1694.1861
Population Analysis
NBO Analysis
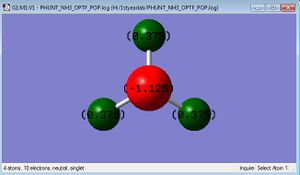
Colour (charge) range: -1.125 to 1.125
Charge on Nitrgoen: -1.125
Charge on Hydrogen: 0.375
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 -0.7062 -0.0240
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 0.7062 0.0239
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756 24(v),16(v),20(v),17(v)
21(v),25(v)
Association Energy
NH3BH3 Optimisation
NH3BH3 was created and optimised by the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: 0
Spin: singlet
Optimisation log file is linked to here
| heading | heading | |
|---|---|---|
| File Name | NH3BH3 opt | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22468960 | a.u. |
| RMS Gradient Norm | 0.00005839 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 5.5651 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 11.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000124 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000660 0.001800 YES
RMS Displacement 0.000304 0.001200 YES
Predicted change in Energy=-1.649603D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.21 -DE/DX = -0.0001 !
! R2 R(2,7) 1.21 -DE/DX = -0.0001 !
! R3 R(3,7) 1.21 -DE/DX = -0.0001 !
! R4 R(4,8) 1.0186 -DE/DX = -0.0001 !
! R5 R(5,8) 1.0186 -DE/DX = -0.0001 !
! R6 R(6,8) 1.0186 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(1,7,2) 113.8754 -DE/DX = 0.0 !
! A2 A(1,7,3) 113.8742 -DE/DX = 0.0 !
! A3 A(1,7,8) 104.5931 -DE/DX = 0.0 !
! A4 A(2,7,3) 113.8743 -DE/DX = 0.0 !
! A5 A(2,7,8) 104.5962 -DE/DX = 0.0 !
! A6 A(3,7,8) 104.6001 -DE/DX = 0.0 !
! A7 A(4,8,5) 107.8698 -DE/DX = 0.0 !
! A8 A(4,8,6) 107.8683 -DE/DX = 0.0 !
! A9 A(4,8,7) 111.0271 -DE/DX = 0.0 !
! A10 A(5,8,6) 107.87 -DE/DX = 0.0 !
! A11 A(5,8,7) 111.0319 -DE/DX = 0.0 !
! A12 A(6,8,7) 111.0285 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.998 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.001 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0012 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0027 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 59.9983 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 180.0005 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 59.9984 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 179.9994 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.9983 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
Link to frequency analysis log file File:NH3BH3 FREQ.LOG
| heading | heading | |
|---|---|---|
| File Name | NH3BH3 freq | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22468959 | a.u. |
| RMS Gradient Norm | 0.00005841 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 5.5651 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 0 minutes 47.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000109 0.000450 YES
RMS Force 0.000058 0.000300 YES
Maximum Displacement 0.000656 0.001800 YES
RMS Displacement 0.000355 0.001200 YES
Predicted change in Energy=-1.748458D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.0011 -0.0008 -0.0007 16.7610 22.0475 39.0806 Low frequencies --- 266.2999 632.2669 638.6816
Calculation
| Molecule | Energy (au) | |
|---|---|---|
| E(NH3) | -56.55776856 | |
| E(BH3) | -26.61532363 | |
| E(NH3BH3) | -83.22468960 | |
Energy difference, ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE= (83.22468960)-(83.17309219) au = 0.05159741 au = 135.47 kJ / mol
This is in good agreement with the literature value of 122.55 kJ/mol.
G. Leroy, M. Sana, C. Wilante , "Evaluation of Bond Energy Terms for the Various Types of Boron-Nitrogen Bonds", Theor Chim Acta, 1983, 85, 1555
Aromaticity Project
Benzene
Benzene Optimisation
Link to optimisation log file is on the [D-space]
Optimisation was carried out with the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: 0
Spin: singlet
| heading | heading | |
|---|---|---|
| File Name | RB benzene optimisation | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -232.25820551 | a.u. |
| RMS Gradient Norm | 0.00009549 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0001 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 2 minutes 11.4 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000212 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000991 0.001800 YES
RMS Displacement 0.000315 0.001200 YES
Predicted change in Energy=-5.157454D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Frequency analysis file is linked to [D-space]
Low frequencies --- -17.3539 -14.6502 -9.7489 -0.0011 -0.0008 -0.0002 Low frequencies --- 413.7962 414.4677 620.8521
The outer limit has just fallen outside the +15 limit, but the calculation should be acceptable.
NBO analysis
The NBO analysis log file is on the [D-space]
Analysis was run with 'Energy', Full NBO and additional keyword "pop=full".
Boratabenzene
Boratabenzene Optimisation
Link to optimisation log file is on the [D-space]
Optimisation was carried out with the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: -1
Spin: singlet
| heading | heading | |
|---|---|---|
| File Name | RB boratabenzene optimisation | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | -1 | |
| Spin | Singlet | |
| E(RB3LYP) | -219.02052985 | a.u. |
| RMS Gradient Norm | 0.00015727 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 2.4865 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 2 minutes 11.4 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000068 0.000300 YES
Maximum Displacement 0.000888 0.001800 YES
RMS Displacement 0.000326 0.001200 YES
Predicted change in Energy=-6.532183D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Frequency analysis file is linked to [D-space]
Low frequencies --- -13.1237 0.0001 0.0008 0.0009 15.0618 18.1745 Low frequencies --- 371.3451 404.2337 565.2515
NBO analysis
The NBO analysis log file is on the [D-space]
Analysis was run with 'Energy', Full NBO and additional keyword "pop=full".
Pyridinium
Pyridinium Optimisation
Link to optimisation log file is on the [D-space]
Optimisation was carried out with the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: +1
Spin: singlet
| heading | heading | |
|---|---|---|
| File Name | RB pyridium optimisation | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| Spin | Singlet | |
| E(RB3LYP) | -248.66807396 | a.u. |
| RMS Gradient Norm | 0.00003911 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.8727 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 4 minutes 3.0 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000064 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000822 0.001800 YES
RMS Displacement 0.000175 0.001200 YES
Predicted change in Energy=-6.915416D-08
Optimization completed.
-- Stationary point found.
Frequency Analysis
Frequency analysis file is linked to [D-space]
Low frequencies --- -7.2125 -0.0010 -0.0007 -0.0007 17.3350 18.5324 Low frequencies --- 392.4554 404.0615 620.4713
NBO analysis
The NBO analysis log file is on the [D-space]
Analysis was run with 'Energy', Full NBO and additional keyword "pop=full".
Borazine
Borazine Optimisation
Link to optimisation log file is on the [D-Space]
Optimisation was carried out with the following method:
Method: Ground State, DFT, Default Spin, B3LYP
Basis set: 6-31G (d, p)
Charge: 0
Spin: singlet
| heading | heading | |
|---|---|---|
| File Name | RB borazine optimisation | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -242.68458488 | a.u. |
| RMS Gradient Norm | 0.00006577 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0002 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 5 minutes 7.3 seconds. | ||
Item Value Threshold Converged?
Maximum Force 0.000093 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000397 0.001800 YES
RMS Displacement 0.000108 0.001200 YES
Predicted change in Energy=-1.074458D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Frequency analysis file is linked to [D-space]
Low frequencies --- -18.0224 -11.8976 -9.6970 -0.0010 -0.0005 0.0013 Low frequencies --- 288.9542 289.5070 404.1692
NBO analysis
The NBO analysis log file is on the [D-space]
Analysis was run with 'Energy', Full NBO and additional keyword "pop=full".
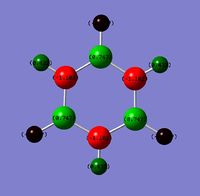
NBO Analysis Summary
| a | benzene | boratabenzene | pyridinium | borazine |
|---|---|---|---|---|
| Charge colour distribution | 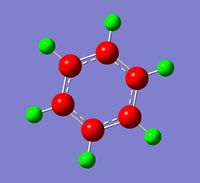 |
 |
 |
|
| Limits | -0.239 to 0.239 | -0.588 to 0.588 | -0.483 to 0.483 | -1.102 to 1.102 |
| Numbers | 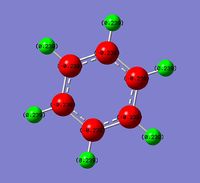 |
 |
 |
|
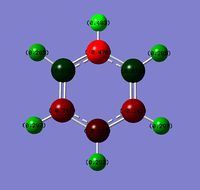
Observing the relative charge distributions, it is seen that there is a uniform charge distribution in benzene.
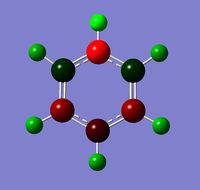
In boratabenzene, the B in C-B is more positive than C, and the B in B-H is more positive than H. This is of course a result of the relative electronegativities of the atoms, with a more electronegative atom attracting the electrons in a covalent bond to itself more strongly than a less electronegative atom, giving a charge distribution.
Boratabenzene has an overall negative charge, by comparing the charge distribution with benzene we can see that this charge is delocalised over the carbons ortho to Boron. In boratabenzene they have -0.588, compared to -0.239 in benzene. The remaining carbons are -0.256, much closer to the benzene values.
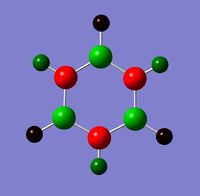
In pyridinium, the H in N-H is more positive than the H in C-H, and as with boratabenzene this can be explained by the relative elctronegativities of the atoms, with nitrogen being more electronegative than carbon.
Pyridinium has an overall positive charge,by comparing the charge distribution with benzene we can see that this charge is delocalised over the carbons ortho to Nitrogen. In pyridinium they have 0.071, compared with -0.239 in benzene. The remaining carbons are -0.241, and as with boratabenzene, closer to the benzene values.
Comparing the charge change in boratabenzene and pyridinium relative to benzene, in boratabenzene the charge has changed by -0.349, whilst in pyridinium the charge has changed by 0.310.
Observing the charge distribution on borazine, we can see that the charges are localised on each B and N, with N clearly being more electronegative than B, giving rise to the observed charge differences (H on N-H more positive than H on B-H).
MOs of Benzene
The pi system is composed of MOs 17, 20-23 and 27 (all fully pi bonding MOs). The HOMO and LUMO are both the pi-system MOs. Huckel's rule states that 4n + 2 = total pi electron occupancy, and that the reactivity of the molecule is most greatly influenced by the pi-orbitals, which as we can see from the MO diagram, the HOMOs are indeed pi-bonding MOs.
The aromaticity in benzene is demonstrated as a result of the overlap of these co-planar pi orbitals.
MO Comparison of studied molecules
In this section 'MOs' have been picked to compare - that is, MOs with similar phase composition, as opposed to trying to compare say, all the MOs that fall on level 8 - which could be problematic as re-ordering of the energy levels can occur, and would make useful comparison difficult. Yet in most cases the MOs do fall on the same energy level, and where a reordering has occurred for a particular molecule, this has been pointed out.
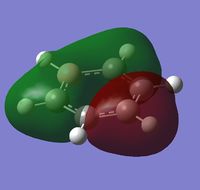
| a | benzene | boratabenzene | pyridinium | borazine |
|---|---|---|---|---|
| Real MO |  |
 |
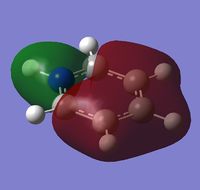 |
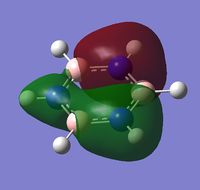
|
| LCAO | 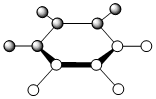 |
 |
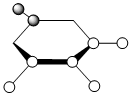 |
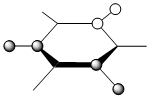
|
| Energy | -0.74006 | -0.46080 | -1.02630 | -0.83513 |
MO Boratabenzene 8 and 9 reordered, so MO '9' has been presented.
| a | benzene | boratabenzene | pyridinium | borazine |
|---|---|---|---|---|
| Real MO |  |
 |
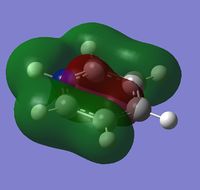 |

|
| LCAO | 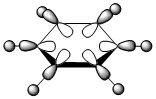 |
 |
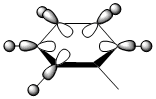 |
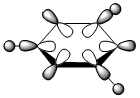
|
| Energy | -0.51794 | -0.28940 | -0.79011 | -0.52454 |
In borazine, MO 12 has been reordered, so MO '10' has been presented.
| a | benzene | boratabenzene | pyridinium | borazine |
|---|---|---|---|---|
| Real MO |  |
 |
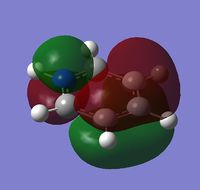 |
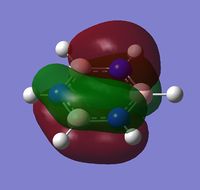
|
| LCAO | 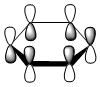 |
 |
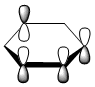 |
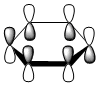
|
| Energy | -0.24692 | -0.03493 | -0.50847 | -0.27591 |
LCAOs contributing to molecules
The contributing LCAOs have been drawn for the chosen MOs as shown above.
MO 8 - each of the MOs are composed of sigma overlaps from s orbitals. Boratabenzene has non-bonding C-H meta to Boron, whilst pyridinium has non-bonding C-H ortho to Nitrogen. Borazine has non-bonding B-H.
MO 12 - the MOs are formed from sigma overlaps of s and p orbitals. There is a pi-bonding contribution, in the creation of the inner ring phase.
MO 20 - the MOs are all composed of fully pi bonding orbitals.
Energy, ordering and degeneracy of the MOs
MO 8 - The energy decreases in the order: boratabenzene < benzene < borazine < pyridinium In boratabenzene MO levels 8 and 9 had been reordered, so in the above tabulation MO 9 has been presented so that comparisons can be drawn between the MOs.
MO 12 - The energy decreases in the order: boratabenzene < benzene < borazine < pyridinium In borazine MO levels 10 and 12 have been reordered, so in the above tabulation MO 10 has been presented so that comparisons can be drawn between the MOs.
MO 20 - The energy decreases in the order: boratabenzene < benzene < borazine < pyridinium
Analysing the MO energy levels, it is observed that there is a significant loss of what were degenerate orbitals in benzene, in both boratabenzene and pyridinium. This results from a loss of symmetry in these molecules.
Looking at the energy decreases, it is seen that pyridinium has the most negative energy, and therefore is the most stabilised. The stabilisation order is hence: pyridinium > borazine > benzene > boratabenzene
This order reflects the natural electronegativity of the substituted atoms; adding the electronegative nitrogen to the ring has lowered the MO levels in energy. Borazine and benzene are seen to be fairly close in the magnitude of their MO energy levels, this likely being as a result of both being neutral, and borazine having an even charge distribution around the molecule due to the equal numbers of B and N, as per benzene experiencing an even charge distribution due to delocalisation.
Similarities and differences between these MOs
MO 8 - each is composed of two phases with a nodal region separating. Benzene and borazine have a nodal region bisecting the molecule through C-C (or N-B) bonds, whilst in boratabenzene the nodal region runs through the carbons meta to Boron, and in pyridinium the nodal region runs through the carbons ortho to Nitrogen. The lobe centered on Boron in boratabenzene is far larger than the lobe centered on Nitrogen in pyridinium. The lobes are of equal size in benzene, whilst in boratabenzene the B-centered lobe is comparatively larger than the other lobe, and in pyridinium the N-centered lobe is comparatively smaller.
MO 12 - The MOs consist of two phases, an inner and outer ring. In benzene, the outer ring is donut shaped with a circular node separating the inner phase. In boratabenzene this outer ring is not fully joined, instead separating into two separate regions that surround the inner phase. In pyridinium, owing to the non-bonding C-H, the outer phase surrounds the inner phase from the side of the nitrogen, existing as one complete phase with a broken region. In borazine, the outer phase exists as six entirely separate regions, with the regions in proximity to boron being far smaller than the regions in proximity to nitrogen.
MO 20 - each is composed of four phases, with two nodes. A node in each case runs through the plane of the ring. The other node is located in a plane analogous to MO 8.
Effect of substitutions on full MO diagram
The substitutions will affect the splitting energy (stabilisation and destabilisation energy), which depends on three quantities, the energy difference between the frontier orbitals, orbital coupling and extent of orbital overlap. Each of these quantities in turn will be affected by which atoms are present in the compound. In this project we have looked at the effect of electropositive and electronegative substitions in a six-membered aromatic ring. Hence, adding either an electropositive or electronegative atom will change those three quantities, and so the HOMO-LUMO gap of the MO diagram will be changed.
Summary
In this project the aromaticity of benzene has been demonstrated through calculation of a Molecular Orbital diagram, and benzene has been compared to its analogues pyridinium, boratabenzene and borazine, by analysing three molecular orbitals from each structure. A quantitative picture has been able to be drawn, rather than a purely qualitative scheme as is possible through LCAO theory alone.