Rep:Mod:rr12104
NH3: optimisation, frequency, NBO
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000071 0.001800 YES
RMS Displacement 0.000034 0.001200 YES
Predicted change in Energy=-5.828500D-10
Optimization completed.
-- Stationary point found.
| Molecule | NH3 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Energy /au | -56.55776863 |
| RMS gradient | 0.00000478 |
| Point group | C3V |
N-H bond distance: 1.018Å
N-H bond angle: 105.743°
NH3 |
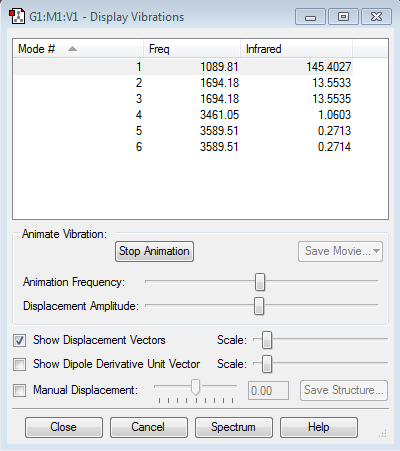
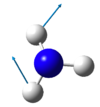
Vibrations
| wavenumber cm-1 |
1092 | 1695 | 1695 | 3458 | 3586 | 3586 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| intensity arbitrary units |
145 | 14 | 14 | 1 | 0 | 0 |
| image | 
|

|

|
|

|
|
How many modes do you expect from the 3N-6 rule?
N=4 so 3N-6 is 6 vibrational modes.
Which modes are degenerate (ie have the same energy)?
modes at 1695 (2 and 3) and 3585 (5 and 6) cm-1 are degenerate
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
bending are 1092 and 1695 cm-1 modes (1,2 and 3), stretching are 3458 and 3586 cm-1 modes (4, 5 and 6), thus stretches are higher in energy than bends
Which mode is highly symmetric?
the totally symmetric mode in at 3458 cm-1 (mode 4)
One mode is known as the "umbrella" mode, which one is this?
the umbrella mode is the mode at 1092 cm-1 (mode 1)
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
The vibration has to cause the dipole has to change to be IR active. Also there are 2 degenerate vibrations which will cause only one peak. Therefore we will see 2 peaks, one very intense one at 1091 cm-1 (mode 1) and one very weak one 1694 cm-1 (degenerate modes 2 and 3).
Charges
Charge on N = -1.125 Charge on H = +0.375
N is more electronegative than H so I would expect to be slightly negatively charged and H to be slightly positively charged.
Reaction and Energy
N2: optimisation, frequency, NBO
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-9.718076D-17
Optimization completed.
-- Stationary point found.
| Molecule | N2 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Energy /au | -109.52412907 |
| RMS gradient | 0.00000001 |
| Point group | DinfH |
N-N bond distance: 1.10550Å
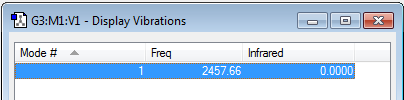
N2 |
| wavenumber cm-1 |
2458 |
|---|---|
| symmetry | SGG or Σg+ |
| intensity arbitrary units |
0 |
| image | to be added |
H2: optimisation, frequency, NBO
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.150392D-13
Optimization completed.
-- Stationary point found.
| Molecule | H2 |
|---|---|
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Energy /au | -1.17853934 |
| RMS gradient | 0.00000017 |
| Point group | DinfH |
H-H bond distance: 0.743Å
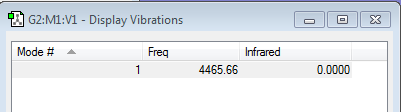
H2 |
| wavenumber cm-1 |
4466 |
|---|---|
| symmetry | SGG or Σg+ |
| intensity arbitrary units |
0 |
| image | to be added |
Structure Comparison
DAYGEP N-N distances in the complex are 1.119(4) and 1.126(4)Å and N-N distance in the computed molecule is 1.106Å The computed gas-phase N-N distance is shorter than the experimental solid state structure N-N distance. In the solid state the N2 molecule is coordinated to a metal, thus some of the bond electron-density will be used to form the M-N2 bond and not in the N-N bond. In the solid state there can also be crystal packing effects, and in this case trans effects from the ligands opposite the N2. The computational result is evaluated in the gas phase, and the N2 is an isolated molecule with no external influences. The bond distance could be changed slightly by using an improved computational method. The error in both the crystal structure and the calculation must also be considered, an error of 0.01 in the computed structure means we must consider the crystal structure and computed molecule distances as equivalent within computational error.
Energies
E(NH3)= -56.55776863au
2*E(NH3)= -113.11553726au
E(N2)= -109.52412907au
E(H2)= -1.17853934au
3*E(H2)= -3.53561802au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.055790170au, -140.90 kJ/mol
The energy decreases from the gases to the ammonia product so the product is more stable.
Detailed CO2 MOs explanation
https://wiki.ch.ic.ac.uk/wiki/index.php?title=Mod:CO2_MO_explanation
10/10 student wikis from 2019
https://wiki.ch.ic.ac.uk/wiki/index.php?title=Mod:Yc16318
https://wiki.ch.ic.ac.uk/wiki/index.php?title=Mod:01349769
https://wiki.ch.ic.ac.uk/wiki/index.php?title=Mod:dialekticnomaterialisticen
HOW TO MARK THE LAB
Note: all google docs/sheets links are view only, make yourself a copy to edit them.
1. Make a google sheet (copy this example one) and add the students names from the main list you will be sent. (If you don't get sent a list you can copy and paste the names from blackboard.)
2. Mark each wiki and give feedback using this template and these model answers added at the end of the students wiki. The first few wikis you should mark with all the markers together to set standards. While marking always ask the other markers if you are unsure on a grade, to make sure you are the same.
3. Type the marks into the google sheet as you go along.
4. Moderation meeting each week with Tricia, use the google sheet to calculate the average for the week and the average for each marker. Grab the links for the best and worst wikis so she can see them.
5. After moderation release the grades via blackboard learn, here is a video showing how: https://www.youtube.com/watch?v=T69nU7FykUs Go to grade centre, needs marking, and filter for the IMM2 lab, then click on a student. Type their grade in the attempt box and click submit. The page will change automatically to the next students submission.