Rep:Mod:psl16
NH3 molecule
Gaussian optimization data
- Molecule name: Ammonia
- Calculation method: RB3LYP
- Basis set: 6-31G(d.p)
- Final energy: -56.55776873 au
- RMS gradient: 0.00000485 au
- Point group: C3V
- Optimized bond length: 1.01798 Angstroms (lit. value: 1.012)[1]
- Optimized H-N-H bond angle: 105.741 degrees (lit. value: 106.7)[1]
Ammonia |
Items table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
The optimisation file is liked to here.
Vibrations
- Expected no. of modes: 6
- Degenerate modes: 2 and 3; 5 and 6
- Modes 1-3 are bending vibrations, modes 4-6 are stretching vibrations
- Mode 4 is highly symmetric
- Mode 1 is the umbrella mode
- You would expect to see 4 bands in an experimental spectrum of ammonia
Charges
The following charges are obtained using NBO.
- Charge on N: -1.125
- Charge on H: 0.375
This is expected as Nitrogen is more electronegative than Hydrogen. Therefore, Nitrogen has a negative charge and Hydrogen has a positive charge.
Born-Haber Process
Nitrogen molecule
Gaussian optimisation data
- Calculation method: RB3LYP
- Basis set: 6-31G(d.p)
- Energy: -109.52412868 au
- RMS gradient: 0.00000060 au
- Point group: Dinf,h
- N-N Bond length: 1.10550 Angstroms (lit. value: 1.0976)[2]
- Vibrational frequency: 2457.33

Nitrogen |
(In the jmol the bond seems like a single bond, so I have included an image as well to show the triple bond)
Items table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
The optimisation file is liked to here.
Hydrogen molecule
Gaussian optimisation data
- Calculation method: RB3LYP
- Basis set: 6-31G(d.p)
- Energy: -1.17853936 au
- RMS gradient: 0.00000017
- Point group: Dinf,h
- Charge: 0
- Spin: singlet
- H-H Bond length: 0.74279 Angstroms (lit.value: 0.74)[3]
- Vibrational frequency: 4465.68
Hydrogen |
Items table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
The optimisation file is liked to here.
Reaction energies
N2 + 3H2 -> 2NH3
- E(NH3) = -56.55776873 au
- 2*E(NH3) = -113.1155375 au
- E(N2) = -109.5241287 au
- E(H2) = -1.17853936 au
- 3*E(H2) = -3.53561808 au
- ΔE = -0.0557907 au = -146.47848 kJ/mol (8 d.p)
The enthalpy change obtained here is quite different from the literature value of -45.7 kJ/mol[4], but this depends on the pressure and temperature at which the reaction is carried out. The reaction is exothermic, so the product is more stable.
ClF3 molecule
Gaussian optimisation data
- Molecule name: Chlorine trifluoride
- Calculation method: RB3LYP
- Basis set: 6-31G(d.p)
- Final energy: -759.46531688au
- RMS gradient: 0.00002465
- Point group: C2V
- Optimized F-Cl bond lengths: 1.72863 (axial F) and 1.65143 (equatorial Fs) Angstroms (lit value: 1.698 and 1.598)[5]
- Optimized F-Cl-F bond angle: 87.140 degrees between ax. and eq. F (lit. value 87.5)[2] and 174.281 (between 2 ax. F)
Chlorine trifluoride |
Items table
Item Value Threshold Converged? Maximum Force 0.000050 0.000450 YES RMS Force 0.000028 0.000300 YES Maximum Displacement 0.000185 0.001800 YES RMS Displacement 0.000134 0.001200 YES
The optimisation file is liked to here.
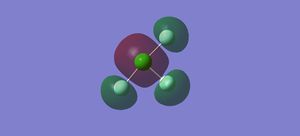

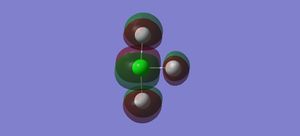
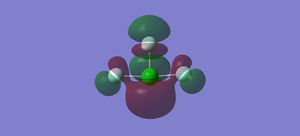
Vibrations
There are 6 vibrational modes, listed in the table below. The first three modes are bending vibrations and the last three are stretching vibrations. There are no degenerate modes. 6 bands are expected in an IR spectrum of the molecule.
| Mode | Frequency | Infrared |
| 1 | 304.79 | 13.5036 |
| 2 | 309.07 | 18.0913 |
| 3 | 401.06 | 0.9480 |
| 4 | 540.78 | 2.7357 |
| 5 | 735.85 | 38.0188 |
| 6 | 752.26 | 370.3594 |
Charges
- Charge on Cl: 1.225
- Charge on Fl: -0.454 on axial Fs and -0.316 on equatorial Fs
This is expected as Fluorine is more electronegative than Chlorine, and therefore draws electron density towards it and has a negative charge.
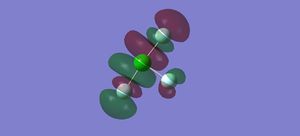
Molecular orbitals
There are 64 molecular orbitals for this molecule in total, of which 22 are occupied. I have included 5 that I find interesting below in order of increasing energy. The first three are occupied, whereas the last two are unoccupied.
In the first molecular orbital, the s orbitals on the Fs are all in phase with each other, but out of phase with the s orbital on the Cl, forming an anti-bonding sigma MO.
In the second molecular orbital, the p orbitals on the axial Fs and the Cl are in phase with each other, forming a bonding pi orbital, but are out of phase with the p orbital on the equatorial Cl, forming an anti-bonding pi orbital.
The third MO is the highest occupied molecular orbital (HOMO), where the pi orbitals on the Fs are in phase with each other, but out of phase with the p orbital on the Cl, forming an anti-bonding pi MO.
The fourth MO is the lowest unoccupied molecular orbital (LUMO), where the p orbital on the Cl is out of phase with the p orbital on all the Fs. Although there is some mixing between the orbitals on the Cl and the axial Fs, they are out of phase where the bonds are located so forms an anti-bonding pi MO.
The fifth MO is the orbital above the LUMO, where all of the p orbitals are out of phase with each other, forming an anti-bonding pi MO.
References
- ↑ 1.0 1.1 CRC Handbook of Chemistry and Physics http://hbcponline.com/faces/contents/ContentsResults.xhtml
- ↑ 2.0 2.1 Greenwood and Earnshaw. Chemistry of the Elements pp412-6
- ↑ Department of Chemistry, University of Texas A&M. http://www.chem.tamu.edu/rgroup/connell/linkfiles/bonds.pdf
- ↑ Modak, J.M. Journal of Science Education. 2002, 7, 9, pp69-77.
- ↑ Smith, D.L. The Journal of Chemical Physics. 1952, 21, 4, pp609-614.